1A5F
 
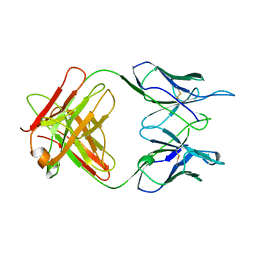 | | FAB FRAGMENT OF A MONOCLONAL ANTI-E-SELECTIN ANTIBODY | | Descriptor: | MONOCLONAL ANTI-E-SELECTIN 7A9 ANTIBODY (HEAVY CHAIN), MONOCLONAL ANTI-E-SELECTIN 7A9 ANTIBODY (LIGHT CHAIN) | | Authors: | Rodriguez-Romero, A, Almog, O, Tordova, M, Randhawa, Z. | | Deposit date: | 1998-02-16 | | Release date: | 1999-04-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Primary and tertiary structures of the Fab fragment of a monoclonal anti-E-selectin 7A9 antibody that inhibits neutrophil attachment to endothelial cells.
J.Biol.Chem., 273, 1998
|
|
1BHN
 
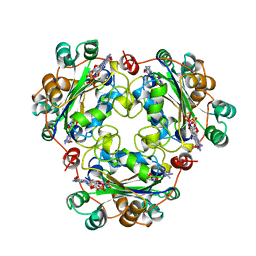 | | NUCLEOSIDE DIPHOSPHATE KINASE ISOFORM A FROM BOVINE RETINA | | Descriptor: | GUANOSINE-3',5'-MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, NUCLEOSIDE DIPHOSPHATE TRANSFERASE | | Authors: | Ladner, J.E, Abdulaev, N.G, Kakuev, D.L, Karaschuk, G.N, Tordova, M, Eisenstein, E, Fujiwara, J.H, Ridge, K.D, Gilliland, G.L. | | Deposit date: | 1998-06-10 | | Release date: | 1999-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structures of two isoforms of nucleoside diphosphate kinase from bovine retina.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BE4
 
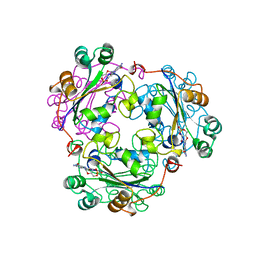 | | NUCLEOSIDE DIPHOSPHATE KINASE ISOFORM B FROM BOVINE RETINA | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, NUCLEOSIDE DIPHOSPHATE TRANSFERASE | | Authors: | Ladner, J.E, Abdulaev, N.G, Kakuev, D.L, Karaschuk, G.N, Tordova, M, Eisenstein, E, Fujiwara, J.H, Ridge, K.D, Gilliland, G.L. | | Deposit date: | 1998-05-19 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nucleoside diphosphate kinase from bovine retina: purification, subcellular localization, molecular cloning, and three-dimensional structure.
Biochemistry, 37, 1998
|
|
5EUG
 
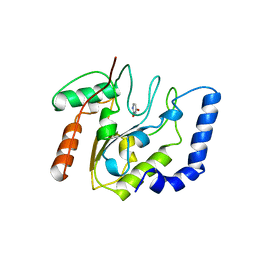 | | CRYSTALLOGRAPHIC AND ENZYMATIC STUDIES OF AN ACTIVE SITE VARIANT H187Q OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE: CRYSTAL STRUCTURES OF MUTANT H187Q AND ITS URACIL COMPLEX | | Descriptor: | PROTEIN (GLYCOSYLASE), URACIL | | Authors: | Xiao, G, Tordova, M, Drohat, A.C, Jagadeesh, J, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-12-27 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
3EUG
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE AND ITS COMPLEXES WITH URACIL AND GLYCEROL: STRUCTURE AND GLYCOSYLASE MECHANISM REVISITED | | Descriptor: | GLYCEROL, PROTEIN (GLYCOSYLASE) | | Authors: | Xiao, G, Tordova, M, Jagadeesh, J, Drohat, A.C, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-10-13 | | Release date: | 1999-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
2EUG
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE AND ITS COMPLEXES WITH URACIL AND GLYCEROL: STRUCTURE AND GLYCOSYLASE MECHANISM REVISITED | | Descriptor: | PROTEIN (GLYCOSYLASE), URACIL | | Authors: | Xiao, G, Tordova, M, Jagadeesh, J, Drohat, A.C, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-10-13 | | Release date: | 1999-10-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
4EUG
 
 | | Crystallographic and Enzymatic Studies of an Active Site Variant H187Q of Escherichia Coli Uracil DNA Glycosylase: Crystal Structures of Mutant H187Q and its Uracil Complex | | Descriptor: | PROTEIN (GLYCOSYLASE) | | Authors: | Xiao, G, Tordova, M, Drohat, A.C, Jagadeesh, J, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-12-27 | | Release date: | 1999-07-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Heteronuclear NMR and crystallographic studies of wild-type and H187Q Escherichia coli uracil DNA glycosylase: electrophilic catalysis of uracil expulsion by a neutral histidine 187.
Biochemistry, 38, 1999
|
|
1FLZ
 
 | | URACIL DNA GLYCOSYLASE WITH UAAP | | Descriptor: | URACIL, URACIL-DNA GLYCOSYLASE | | Authors: | Werner, R.M, Jiang, Y.L, Gordley, R.G, Jagadeesh, G.J, Ladner, J.E, Xiao, G, Tordova, M, Gilliland, G.L, Stivers, J.T. | | Deposit date: | 2000-08-15 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stressing-out DNA? The contribution of serine-phosphodiester interactions in catalysis by uracil DNA glycosylase.
Biochemistry, 39, 2000
|
|
1EUG
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE AND ITS COMPLEXES WITH URACIL AND GLYCEROL: STRUCTURE AND GLYCOSYLASE MECHANISM REVISITED | | Descriptor: | PROTEIN (GLYCOSYLASE) | | Authors: | Xiao, G, Tordova, M, Jagadeesh, J, Drohat, A.C, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-10-12 | | Release date: | 1999-10-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
1HW5
 
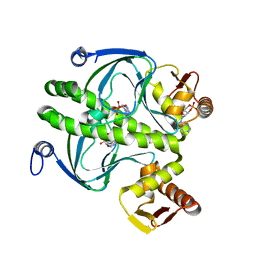 | | THE CAP/CRP VARIANT T127L/S128A | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR | | Authors: | Chu, S.Y, Tordova, M, Gilliland, G.L, Gorshkova, I, Shi, Y. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The structure of the T127L/S128A mutant of cAMP receptor protein facilitates promoter site binding
J.Biol.Chem., 276, 2001
|
|
1I2K
 
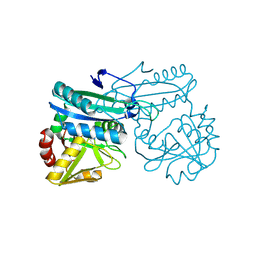 | | AMINODEOXYCHORISMATE LYASE FROM ESCHERICHIA COLI | | Descriptor: | 4-AMINO-4-DEOXYCHORISMATE LYASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jensen, P.Y, Parsons, J.F, Fisher, K.E, Pachikara, A.S, Tordova, M, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-02-09 | | Release date: | 2003-09-02 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure and Mechanism of Escherichia coli Aminodeoxychorismate Lyase
To be Published
|
|
1I2L
 
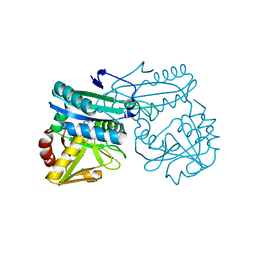 | | DEOXYCHORISMATE LYASE FROM ESCHERICHIA COLI WITH INHIBITOR | | Descriptor: | 4-AMINO-4-DEOXYCHORISMATE LYASE, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Jensen, P.Y, Parsons, J.F, Fisher, K.E, Pachikara, A.S, Tordova, M, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-02-09 | | Release date: | 2003-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of Escherichia coli Aminodeoxychorismate Lyase
To be Published
|
|
1J9A
 
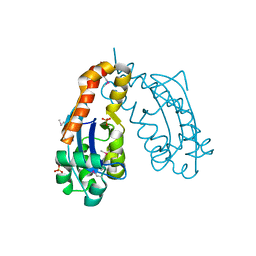 | | OLIGORIBONUCLEASE | | Descriptor: | OLIGORIBONUCLEASE, SULFATE ION | | Authors: | Bonander, N, Tordova, M, Ladner, J.E, Eisenstein, E, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-24 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of Haemophilus Influenzae HI1715, an Oligoribonuclease
To be Published
|
|
1JOV
 
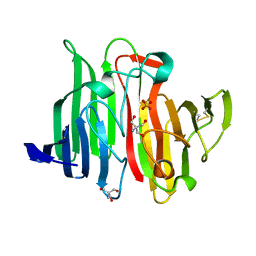 | | Crystal Structure Analysis of HI1317 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HI1317, SULFATE ION | | Authors: | Bonander, N, Tordova, M, Howard, A.J, Eisenstein, E, Gilliland, G, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-31 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal 1.57-A Crystal Structure of HI1317
TO BE PUBLISHED
|
|
1JOS
 
 | | Ribosome Binding Factor A(rbfA) | | Descriptor: | RIBOSOME-BINDING FACTOR A | | Authors: | Bonander, N, Tordova, M, Howard, A.J, Eisenstein, E, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-30 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7-A Crystal Structure of HI1288 - Ribosome Binding Factor A (rbfA), a Cold Response Protein
To be Published
|
|
1PGT
 
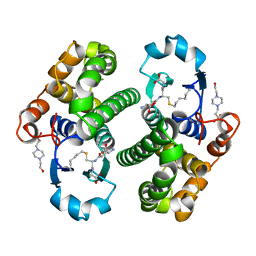 | |
2PGT
 
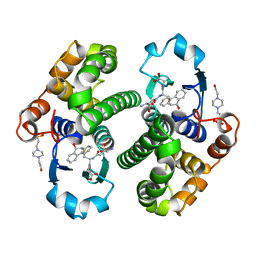 | | CRYSTAL STRUCTURE OF HUMAN GLUTATHIONE S-TRANSFERASE P1-1[V104] COMPLEXED WITH (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, ... | | Authors: | Ji, X. | | Deposit date: | 1997-02-17 | | Release date: | 1997-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the xenobiotic substrate-binding site and location of a potential non-substrate-binding site in a class pi glutathione S-transferase.
Biochemistry, 36, 1997
|
|
1SUA
 
 | | SUBTILISIN BPN' | | Descriptor: | SUBTILISIN BPN', TETRAPEPTIDE ALA-LEU-ALA-LEU | | Authors: | Almog, O, Gilliland, G.L. | | Deposit date: | 1997-01-14 | | Release date: | 1998-01-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of calcium-independent subtilisin BPN' with restored thermal stability folded without the prodomain.
Proteins, 31, 1998
|
|
1XCA
 
 | | APO-CELLULAR RETINOIC ACID BINDING PROTEIN II | | Descriptor: | CELLULAR RETINOIC ACID BINDING PROTEIN TYPE II | | Authors: | Chen, X, Ji, X. | | Deposit date: | 1996-12-31 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of apo-cellular retinoic acid-binding protein type II (R111M) suggests a mechanism of ligand entry.
J.Mol.Biol., 278, 1998
|
|
3PGT
 
 | | CRYSTAL STRUCTURE OF HGSTP1-1[I104] COMPLEXED WITH THE GSH CONJUGATE OF (+)-ANTI-BPDE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-4-[1-(CARBOXYMETHYL-CARBAMOYL)-2-(9-HYDROXY-7,8-DIOXO-7,8,9,10-TETRAHYDRO-BENZO[DEF]CHRYSEN-10-YLSULFANYL)-ETHYLCARBAMOYL]-BUTYRIC ACID, PROTEIN (GLUTATHIONE S-TRANSFERASE), ... | | Authors: | Ji, X, Xiao, B. | | Deposit date: | 1999-03-22 | | Release date: | 1999-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure and function of residue 104 and water molecules in the xenobiotic substrate-binding site in human glutathione S-transferase P1-1.
Biochemistry, 38, 1999
|
|
4PGT
 
 | | CRYSTAL STRUCTURE OF HGSTP1-1[V104] COMPLEXED WITH THE GSH CONJUGATE OF (+)-ANTI-BPDE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-4-[1-(CARBOXYMETHYL-CARBAMOYL)-2-(9-HYDROXY-7,8-DIOXO-7,8,9,10-TETRAHYDRO-BENZO[DEF]CHRYSEN-10-YLSULFANYL)-ETHYLCARBAMOYL]-BUTYRIC ACID, PROTEIN (GLUTATHIONE S-TRANSFERASE), ... | | Authors: | Ji, X, Blaszczyk, J. | | Deposit date: | 1999-03-22 | | Release date: | 1999-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of residue 104 and water molecules in the xenobiotic substrate-binding site in human glutathione S-transferase P1-1.
Biochemistry, 38, 1999
|
|
1DBF
 
 | |
1DSF
 
 | |
1FL9
 
 | | THE YJEE PROTEIN | | Descriptor: | HYPOTHETICAL PROTEIN HI0065, MERCURY (II) ION | | Authors: | Teplyakov, A, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2000-08-13 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the YjeE protein from Haemophilus influenzae: a putative Atpase involved in cell wall synthesis
Proteins, 48, 2002
|
|
1HTW
 
 | |
