1CBU
 
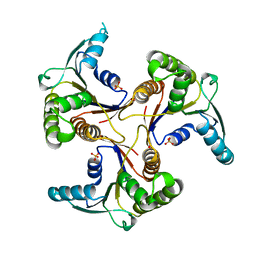 | | ADENOSYLCOBINAMIDE KINASE/ADENOSYLCOBINAMIDE PHOSPHATE GUANYLYLTRANSFERASE (COBU) FROM SALMONELLA TYPHIMURIUM | | Descriptor: | ADENOSYLCOBINAMIDE KINASE/ADENOSYLCOBINAMIDE PHOSPHATE GUANYLYLTRANSFERASE, SULFATE ION | | Authors: | Thompson, T.B, Thomas, M.G, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 1998-03-12 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of adenosylcobinamide kinase/adenosylcobinamide phosphate guanylyltransferase from Salmonella typhimurium determined to 2.3 A resolution,.
Biochemistry, 37, 1998
|
|
1C9K
 
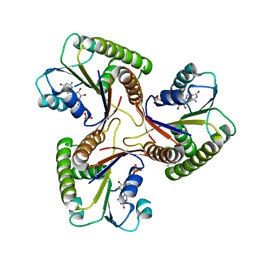 | | THE THREE DIMENSIONAL STRUCTURE OF ADENOSYLCOBINAMIDE KINASE/ ADENOSYLCOBINAMIDE PHOSPHATE GUALYLYLTRANSFERASE (COBU) COMPLEXED WITH GMP: EVIDENCE FOR A SUBSTRATE INDUCED TRANSFERASE ACTIVE SITE | | Descriptor: | ADENOSYLCOBINAMIDE KINASE, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thompson, T.B, Thomas, M.G, Esclante-Semerena, J.C, Rayment, I. | | Deposit date: | 1999-08-02 | | Release date: | 1999-08-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of adenosylcobinamide kinase/adenosylcobinamide phosphate guanylyltransferase (CobU) complexed with GMP: evidence for a substrate-induced transferase active site.
Biochemistry, 38, 1999
|
|
4Q29
 
 | | Ensemble Refinement of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 protein | | Authors: | Wang, F, Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Miller, M.D, Thomas, M.G, Joachimiak, A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-07 | | Last modified: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
4QBU
 
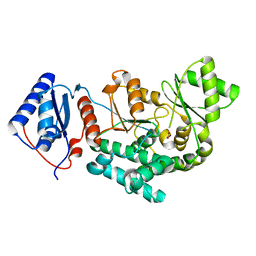 | | Structure of the Acyl Transferase domain of ZmaA | | Descriptor: | FORMIC ACID, ZmaA | | Authors: | Dyer, D.H, Kevany, B.M, Thomas, M.G, Forest, K.T. | | Deposit date: | 2014-05-08 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Polyketide Synthase Acyltransferase Domain Structure Suggests a Recognition Mechanism for Its Hydroxymalonyl-Acyl Carrier Protein Substrate.
Plos One, 9, 2014
|
|
4MV2
 
 | | Crystal structure of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 | | Authors: | Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Thomas, M.G, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-02 | | Last modified: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
5CQF
 
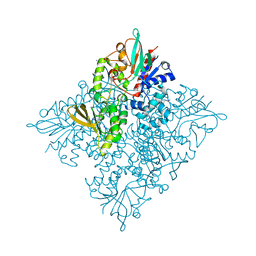 | | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae | | Descriptor: | IODIDE ION, L-lysine 6-monooxygenase | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Weerth, R.S, Cao, H, Yennamalli, R, Phillips Jr, G.N, Thomas, M.G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae
To Be Published
|
|
2WBP
 
 | | Crystal structure of VioC in complex with Fe(II), (2S,3S)- hydroxyarginine, and succinate | | Descriptor: | (2S,3S)-3-HYDROXYARGININE, ARGININE, FE (II) ION, ... | | Authors: | Helmetag, V, Samel, S.A, Thomas, M.G, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2009-03-02 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structural Basis for the Erythro-Stereospecificity of the L-Arginine Oxygenase Vioc in Viomycin Biosynthesis.
FEBS J., 276, 2009
|
|
2WBQ
 
 | | Crystal structure of VioC in complex with (2S,3S)-hydroxyarginine | | Descriptor: | (2S,3S)-3-HYDROXYARGININE, ACETATE ION, L-ARGININE BETA-HYDROXYLASE | | Authors: | Helmetag, V, Samel, S.A, Thomas, M.G, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2009-03-02 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Basis for the Erythro-Stereospecificity of the L-Arginine Oxygenase Vioc in Viomycin Biosynthesis.
FEBS J., 276, 2009
|
|
2WBO
 
 | | Crystal structure of VioC in complex with L-arginine | | Descriptor: | ARGININE, FE (II) ION, L(+)-TARTARIC ACID, ... | | Authors: | Helmetag, V, Samel, S.A, Thomas, M.G, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2009-03-02 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Basis for the Erythro-Stereospecificity of the L-Arginine Oxygenase Vioc in Viomycin Biosynthesis.
FEBS J., 276, 2009
|
|
6SWX
 
 | | Leishmania major methionyl-tRNA synthetase in complex with an allosteric inhibitor | | Descriptor: | METHIONINE, Putative methionyl-tRNA synthetase, methyl 2-[[6-[[3,4-bis(fluoranyl)phenyl]amino]-1-methyl-pyrazolo[3,4-d]pyrimidin-4-yl]amino]ethanoate | | Authors: | Robinson, D.A, Torrie, L.S, Shepherd, S.M, De Rycker, M, Thomas, M.G, Wyatt, P.G. | | Deposit date: | 2019-09-24 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of an Allosteric Binding Site in Kinetoplastid Methionyl-tRNA Synthetase.
Acs Infect Dis., 6, 2020
|
|
8OLU
 
 | |
6CBA
 
 | | Ethylene forming enzyme in complex with manganese, 2-oxoglutarate and canavanine | | Descriptor: | 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, L-CANAVANINE, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2018-02-02 | | Release date: | 2019-02-06 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structural, Spectroscopic, and Computational Insights from Canavanine-Bound and Two Catalytically Compromised Variants of the Ethylene-Forming Enzyme.
Biochemistry, 2024
|
|
6CF3
 
 | | Ethylene forming enzyme Y306A variant in complex with manganese and 2-oxoglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2018-02-13 | | Release date: | 2019-02-13 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural, Spectroscopic, and Computational Insights from Canavanine-Bound and Two Catalytically Compromised Variants of the Ethylene-Forming Enzyme.
Biochemistry, 2024
|
|
8UC2
 
 | | Ethylene forming enzyme (EFE) R171A variant in complex with nickel and Benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, BENZOIC ACID, ... | | Authors: | Chatterjee, S, Rankin, J.A, Hu, J, Hausinger, R. | | Deposit date: | 2023-09-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, Spectroscopic, and Computational Insights from Canavanine-Bound and Two Catalytically Compromised Variants of the Ethylene-Forming Enzyme.
Biochemistry, 63, 2024
|
|
