1C4L
 
 | | SOLUTION STRUCTURE OF AN RNA DUPLEX INCLUDING A C-U BASE-PAIR | | Descriptor: | RNA (5'-R(*CP*CP*UP*GP*CP*GP*UP*CP*G)-3'), RNA (5'-R(*CP*GP*AP*CP*UP*CP*AP*GP*G)-3') | | Authors: | Tanaka, Y, Kojima, C, Yamazaki, T, Kodama, T.S, Yasuno, K, Miyashita, S, Ono, A.M, Ono, A.S, Kainosho, M, Kyogoku, Y. | | Deposit date: | 1999-08-30 | | Release date: | 2000-08-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA duplex including a C-U base pair.
Biochemistry, 39, 2000
|
|
413D
 
 | | A'-FORM RNA DOUBLE HELIX IN THE SINGLE CRYSTAL STRUCTURE OF R(UGAGCUUCGGCUC) | | Descriptor: | RNA (5'-R(*UP*GP*AP*GP*CP*UP*UP*CP*GP*GP*CP*UP*C)-3') | | Authors: | Tanaka, Y, Fujii, S, Hiroaki, H, Sakata, T, Tanaka, T, Uesugi, S, Tomita, K.-I, Kyogoku, Y. | | Deposit date: | 1998-07-10 | | Release date: | 1998-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A'-form RNA double helix in the single crystal structure of r(UGAGCUUCGGCUC).
Nucleic Acids Res., 27, 1999
|
|
6R83
 
 | | CryoEM structure and molecular model of squid hemocyanin (Todarodes pacificus , TpH) | | Descriptor: | Hemocyanin subunit 1 | | Authors: | Tanaka, Y, Kato, S, Stabrin, M, Raunser, S, Matsui, T, Gatsogiannis, C. | | Deposit date: | 2019-03-31 | | Release date: | 2019-05-08 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Cryo-EM reveals the asymmetric assembly of squid hemocyanin.
Iucrj, 6, 2019
|
|
1J2V
 
 | | Crystal Structure of CutA1 from Pyrococcus Horikoshii | | Descriptor: | 102AA long hypothetical periplasmic divalent cation tolerance protein CUTA | | Authors: | Tanaka, Y, Sakai, N, Yasutake, Y, Yao, M, Tsumoto, K, Kumagai, I, Tanaka, I. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural implications for heavy metal-induced reversible assembly and aggregation of a protein: the case of Pyrococcus horikoshii CutA.
Febs Lett., 556, 2004
|
|
1J08
 
 | | Crystal structure of glutaredoxin-like protein from Pyrococcus horikoshii | | Descriptor: | glutaredoxin-like protein | | Authors: | Tanaka, Y, Tanabe, E, Tsumoto, K, Kumagai, I, Yao, M, Tanaka, I. | | Deposit date: | 2002-11-11 | | Release date: | 2003-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein disulfide isomerase from hyperthermophile as an additives of refolding of an immunoglobulin-folded protein
To be Published
|
|
5AWW
 
 | | Precise Resting State of Thermus thermophilus SecYEG | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Protein translocase subunit SecE, Protein translocase subunit SecY, ... | | Authors: | Tanaka, Y, Sugano, Y, Takemoto, M, Kusakizako, T, Kumazaki, K, Ishitani, R, Nureki, O, Tsukazaki, T. | | Deposit date: | 2015-07-10 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.724 Å) | | Cite: | Crystal Structures of SecYEG in Lipidic Cubic Phase Elucidate a Precise Resting and a Peptide-Bound State.
Cell Rep, 13, 2015
|
|
5CH4
 
 | | Peptide-Bound State of Thermus thermophilus SecYEG | | Descriptor: | Protein translocase subunit SecE, Protein translocase subunit SecY, Putative preprotein translocase, ... | | Authors: | Tanaka, Y, Sugano, Y, Takemoto, M, Kusakizako, T, Kumazaki, K, Ishitani, R, Nureki, O, Tsukazaki, T. | | Deposit date: | 2015-07-10 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Crystal Structures of SecYEG in Lipidic Cubic Phase Elucidate a Precise Resting and a Peptide-Bound State.
Cell Rep, 13, 2015
|
|
1UKU
 
 | | Crystal Structure of Pyrococcus horikoshii CutA1 Complexed with Cu2+ | | Descriptor: | COPPER (II) ION, periplasmic divalent cation tolerance protein CutA | | Authors: | Tanaka, Y, Yasutake, Y, Yao, M, Sakai, N, Tanaka, I, Tsumoto, K, Kumagai, I. | | Deposit date: | 2003-09-01 | | Release date: | 2004-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural implications for heavy metal-induced reversible assembly and aggregation of a protein: the case of Pyrococcus horikoshii CutA.
Febs Lett., 556, 2004
|
|
1UMJ
 
 | | Crystal structure of Pyrococcus horikoshii CutA in the presence of 3M guanidine hydrochloride | | Descriptor: | GUANIDINE, periplasmic divalent cation tolerance protein CutA | | Authors: | Tanaka, Y, Tsumoto, K, Yasutake, Y, Sakai, N, Yao, M, Tanaka, I, Kumagai, I. | | Deposit date: | 2003-10-02 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural evidence for guanidine-protein side chain interactions: crystal structure of CutA from Pyrococcus horikoshii in 3M guanidine hydrochloride
Biochem.Biophys.Res.Commun., 323, 2004
|
|
1VBF
 
 | | Crystal structure of protein L-isoaspartate O-methyltransferase homologue from Sulfolobus tokodaii | | Descriptor: | 231aa long hypothetical protein-L-isoaspartate O-methyltransferase | | Authors: | Tanaka, Y, Tsumoto, K, Yasutake, Y, Umetsu, M, Yao, M, Tanaka, I, Fukada, H, Kumagai, I. | | Deposit date: | 2004-02-25 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | How Oligomerization Contributes to the Thermostability of an Archaeon Protein: PROTEIN L-ISOASPARTYL-O-METHYLTRANSFERASE FROM SULFOLOBUS TOKODAII
J.Biol.Chem., 279, 2004
|
|
1WV3
 
 | |
2DSO
 
 | | Crystal structure of D138N mutant of Drp35, a 35kDa drug responsive protein from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Drp35, GLYCEROL | | Authors: | Tanaka, Y, Ohki, Y, Morikawa, K, Yao, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-07-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mutational Analyses of Drp35 from Staphylococcus aureus: A POSSIBLE MECHANISM FOR ITS LACTONASE ACTIVITY
J.Biol.Chem., 282, 2007
|
|
2DG0
 
 | | Crystal structure of Drp35, a 35kDa drug responsive protein from Staphylococcus aureus | | Descriptor: | DrP35 | | Authors: | Tanaka, Y, Ohki, Y, Morikawa, K, Yao, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-03-07 | | Release date: | 2006-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mutational Analyses of Drp35 from Staphylococcus aureus: A POSSIBLE MECHANISM FOR ITS LACTONASE ACTIVITY
J.Biol.Chem., 282, 2007
|
|
2DG1
 
 | | Crystal structure of Drp35, a 35kDa drug responsive protein from Staphylococcus aureus, complexed with Ca2+ | | Descriptor: | CALCIUM ION, DrP35, GLYCEROL | | Authors: | Tanaka, Y, Ohki, Y, Morikawa, K, Yao, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-03-07 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural and Mutational Analyses of Drp35 from Staphylococcus aureus: A POSSIBLE MECHANISM FOR ITS LACTONASE ACTIVITY
J.Biol.Chem., 282, 2007
|
|
5X3J
 
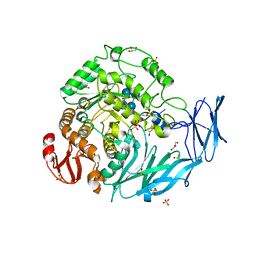 | | Kfla1895 D451A mutant in complex with cyclobis-(1->6)-alpha-nigerosyl | | Descriptor: | Cyclic alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose, GLYCEROL, Glycoside hydrolase family 31, ... | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycoside hydrolase mutant in complex with substrate
To Be Published
|
|
5X3I
 
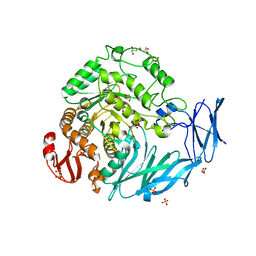 | | Kfla1895 D451A mutant | | Descriptor: | GLYCEROL, Glycoside hydrolase family 31, SULFATE ION | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glycoside hydrolase mutant
To Be Published
|
|
2DGJ
 
 | | Crystal structure of EbhA (756-1003 domain) from Staphylococcus aureus | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Tanaka, Y, Yao, M, Kuroda, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-03-14 | | Release date: | 2007-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A helical string of alternately connected three-helix bundles for the cell wall-associated adhesion protein Ebh from Staphylococcus aureus
Structure, 16, 2008
|
|
1HLV
 
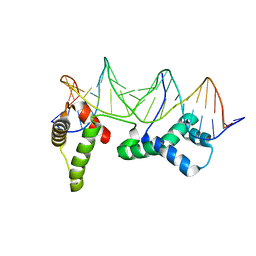 | | CRYSTAL STRUCTURE OF CENP-B(1-129) COMPLEXED WITH THE CENP-B BOX DNA | | Descriptor: | CENP-B BOX DNA, MAJOR CENTROMERE AUTOANTIGEN B | | Authors: | Tanaka, Y, Nureki, O, Kurumizaka, H, Fukai, S, Kawaguchi, S, Ikuta, M, Iwahara, J, Okazaki, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-12-04 | | Release date: | 2002-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the CENP-B protein-DNA complex: the DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA.
EMBO J., 20, 2001
|
|
1WU7
 
 | | Crystal structure of histidyl-tRNA synthetase from Thermoplasma acidophilum | | Descriptor: | Histidyl-tRNA synthetase | | Authors: | Tanaka, Y, Sakai, N, Yao, M, Watanabe, N, Tamura, T, Tanaka, I. | | Deposit date: | 2004-12-01 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of histidyl-tRNA synthetase from Thermoplasma acidophilum
To be Published
|
|
2DGD
 
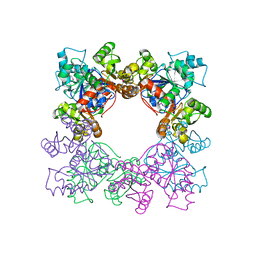 | | Crystal structure of ST0656, a function unknown protein from Sulfolobus tokodaii | | Descriptor: | 223aa long hypothetical arylmalonate decarboxylase, GLYCEROL | | Authors: | Tanaka, Y, Sasaki, T, Tanabe, E, Yao, M, Tanaka, I, Kumagai, I, Tsumoto, K. | | Deposit date: | 2006-03-10 | | Release date: | 2007-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of ST0656, a function unknown protein from Sulfolobus tokodaii
To be Published
|
|
6LEP
 
 | | Crystal structure of thiosulfate transporter YeeE inactive mutant - C91A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Sulf_transp domain-containing protein, THIOSULFATE | | Authors: | Tanaka, Y, Tsukazaki, T, Yoshikaie, K, Sugano, Y, Takeuchi, A, Uchino, S. | | Deposit date: | 2019-11-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a YeeE/YedE family protein engaged in thiosulfate uptake.
Sci Adv, 6, 2020
|
|
6LEO
 
 | | Crystal structure of thiosulfate transporter YeeE from Spirochaeta thermophila | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Sulf_transp domain-containing protein, THIOSULFATE | | Authors: | Tanaka, Y, Tsukazaki, T, Yoshikaie, K, Takeuchi, A, Uchino, S, Sugano, Y. | | Deposit date: | 2019-11-26 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of a YeeE/YedE family protein engaged in thiosulfate uptake.
Sci Adv, 6, 2020
|
|
7DQK
 
 | | A nicotine MATE transporter, Nicotiana tabacum MATE2 (NtMATE2) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanaka, Y, Tsukazaki, T, Sasaki, A, Iwaki, S. | | Deposit date: | 2020-12-24 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of a nicotine MATE transporter provide insight into its mechanism of substrate transport.
Febs Lett., 595, 2021
|
|
5X3K
 
 | | Kfla1895 D451A mutant in complex with isomaltose | | Descriptor: | GLYCEROL, Glycoside hydrolase family 31, SULFATE ION, ... | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glycoside hydrolase mutant in complex with product
To Be Published
|
|
5XJJ
 
 | | Crystal structure of a MATE family protein | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, multi drug efflux transporter | | Authors: | Tanaka, Y, Tsukazaki, T, Iwaki, S. | | Deposit date: | 2017-05-02 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Plant Multidrug and Toxic Compound Extrusion Family Protein
Structure, 25, 2017
|
|
