5GQD
 
 | | Crystal structure of covalent glycosyl-enzyme intermediate of xylanase mutant (T82A, N127S, and E128H) from Streptomyces olivaceoviridis E-86 | | Descriptor: | Beta-xylanase, GLYCEROL, beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Suzuki, R, Fujimoto, Z, Kaneko, S, Kuno, A. | | Deposit date: | 2016-08-07 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Azidolysis by the Formation of Stable Ser-His Catalytic Dyad in a Glycoside Hydrolase Family 10 Xylanase Mutant
J.Appl.Glyosci., 65, 2019
|
|
5GQE
 
 | | Crystal structure of michaelis complex of xylanase mutant (T82A, N127S, and E128H) from Streptomyces olivaceoviridis E-86 | | Descriptor: | Beta-xylanase, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Suzuki, R, Fujimoto, Z, Kaneko, S, Kuno, A. | | Deposit date: | 2016-08-07 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Azidolysis by the Formation of Stable Ser-His Catalytic Dyad in a Glycoside Hydrolase Family 10 Xylanase Mutant
J.Appl.Glyosci., 65, 2019
|
|
2RQF
 
 | | Solution structure of juvenile hormone binding protein from silkworm in complex with JH III | | Descriptor: | Hemolymph juvenile hormone binding protein, methyl (2E,6E)-9-[(2R)-3,3-dimethyloxiran-2-yl]-3,7-dimethylnona-2,6-dienoate | | Authors: | Suzuki, R, Fujimoto, Z, Shiotsuki, T, Momma, M, Tase, A, Yamazaki, T. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-05 | | Last modified: | 2013-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural mechanism of JH delivery in hemolymph by JHBP of silkworm, Bombyx mori
Sci Rep, 1, 2011
|
|
2RNN
 
 | |
2RNO
 
 | |
1IX5
 
 | | Solution structure of the Methanococcus thermolithotrophicus FKBP | | Descriptor: | FKBP | | Authors: | Suzuki, R, Nagata, K, Kawakami, M, Nemoto, N, Furutani, M, Adachi, K, Maruyama, T, Tanokura, M. | | Deposit date: | 2002-06-12 | | Release date: | 2003-06-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional Solution Structure of an Archaeal FKBP with a Dual Function of Peptidyl Prolyl cis-trans Isomerase and Chaperone-like Activities
J.MOL.BIOL., 328, 2003
|
|
2D1Z
 
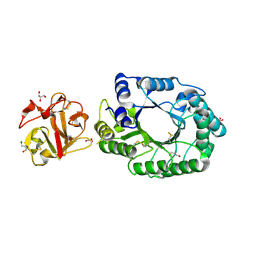 | | Crystal structure of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D20
 
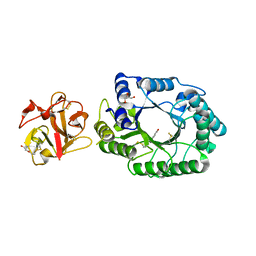 | | Crystal structure of michaelis complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D23
 
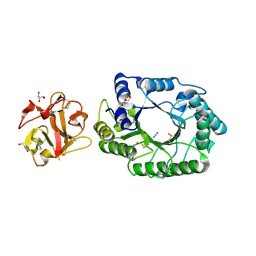 | | Crystal structure of EP complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | AZIDE ION, ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
3WNP
 
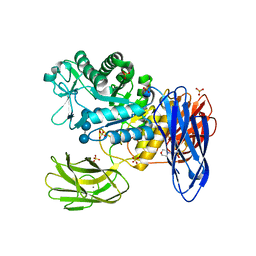 | | D308A, F268V, D469Y, A513V, and Y515S quintuple mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltoundecaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, R, Suzuki, N, Fujimoto, Z, Momma, M, Kimura, K, Kitamura, S, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular engineering of cycloisomaltooligosaccharide glucanotransferase from Bacillus circulans T-3040: structural determinants for the reaction product size and reactivity.
Biochem.J., 467, 2015
|
|
2D24
 
 | | Crystal structure of ES complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D22
 
 | | Crystal structure of covalent glycosyl-enzyme intermediate of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2ZQO
 
 | | Crystal structure of the earthworm R-type lectin C-half in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 29-kDa galactose-binding lectin, CADMIUM ION, ... | | Authors: | Suzuki, R, Kuno, A, Hasegawa, T, Hirabayashi, J, Kasai, K, Momma, M, Fujimoto, Z. | | Deposit date: | 2008-08-13 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sugar-complex structures of the C-half domain of the galactose-binding lectin EW29 from the earthworm Lumbricus terrestris
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZQN
 
 | | Crystal structure of the earthworm R-type lectin C-half in complex with Lactose | | Descriptor: | 29-kDa galactose-binding lectin, IMIDAZOLE, PHOSPHATE ION, ... | | Authors: | Suzuki, R, Kuno, A, Hasegawa, T, Hirabayashi, J, Kasai, K, Momma, M, Fujimoto, Z. | | Deposit date: | 2008-08-13 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sugar-complex structures of the C-half domain of the galactose-binding lectin EW29 from the earthworm Lumbricus terrestris
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2Z8F
 
 | | The galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum in complex with lacto-N-tetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Galacto-N-biose/lacto-N-biose I transporter substrate-binding protein, SODIUM ION, ... | | Authors: | Suzuki, R, Wada, J, Katayama, T, Fushinobu, S. | | Deposit date: | 2007-09-05 | | Release date: | 2008-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I.
J.Biol.Chem., 283, 2008
|
|
3AHI
 
 | | H320A mutant of Phosphoketolase from Bifidobacterium Breve complexed with acetyl thiamine diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-ACETYL-THIAMINE DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
3AHJ
 
 | | H553A mutant of Phosphoketolase from Bifidobacterium Breve | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
3AHC
 
 | | Resting form of Phosphoketolase from Bifidobacterium Breve | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NONAETHYLENE GLYCOL, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
3AHE
 
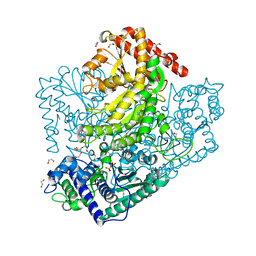 | | Phosphoketolase from Bifidobacterium Breve complexed with dihydroxyethyl thiamine diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-[(4-AMINO-2-METHYL-5-PYRIMIDINYL)METHYL]-2-(1,2-DIHYDROXYETHYL)-4-METHYL-1,3-THIAZOL-3-IUM-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
2Z8E
 
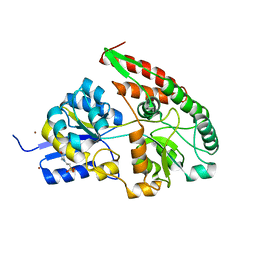 | | The galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum in complex with galacto-N-biose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Galacto-N-biose/lacto-N-biose I transporter substrate-binding protein, ZINC ION, ... | | Authors: | Suzuki, R, Wada, J, Katayama, T, Fushinobu, S. | | Deposit date: | 2007-09-05 | | Release date: | 2008-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I.
J.Biol.Chem., 283, 2008
|
|
3AHD
 
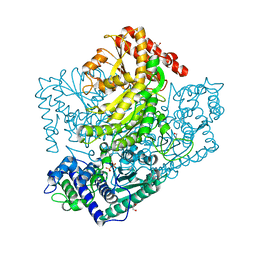 | | Phosphoketolase from Bifidobacterium Breve complexed with 2-acetyl-thiamine diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-ACETYL-THIAMINE DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
3AHF
 
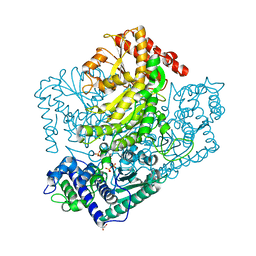 | | Phosphoketolase from Bifidobacterium Breve complexed with inorganic phosphate | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
3AHH
 
 | | H142A mutant of Phosphoketolase from Bifidobacterium Breve complexed with acetyl thiamine diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-ACETYL-THIAMINE DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
2Z8D
 
 | | The galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum in complex with lacto-N-biose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Galacto-N-biose/lacto-N-biose I transporter substrate-binding protein, ZINC ION, ... | | Authors: | Suzuki, R, Wada, J, Katayama, T, Fushinobu, S. | | Deposit date: | 2007-09-05 | | Release date: | 2008-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I.
J.Biol.Chem., 283, 2008
|
|
3AHG
 
 | | H64A mutant of Phosphoketolase from Bifidobacterium Breve complexed with a tricyclic ring form of thiamine diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-[(9aR)-2,7-dimethyl-9a,10-dihydro-5H-pyrimido[4,5-d][1,3]thiazolo[3,2-a]pyrimidin-8-yl]ethyl trihydrogen diphosphate, MAGNESIUM ION, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
