5WQL
 
 | |
2M5V
 
 | |
6V4U
 
 | | Cryo-EM structure of SMCR8-C9orf72-WDR41 complex | | Descriptor: | Guanine nucleotide exchange C9orf72, Guanine nucleotide exchange protein SMCR8, WD repeat-containing protein 41 | | Authors: | Su, M.Y, Hurley, J.H. | | Deposit date: | 2019-12-01 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the C9orf72 ARF GAP complex that is haploinsufficient in ALS and FTD.
Nature, 585, 2020
|
|
8WQR
 
 | | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation | | Descriptor: | Activating molecule in BECN1-regulated autophagy protein 1, DNA damage-binding protein 1 | | Authors: | Liu, M, Wang, Y, Su, M.Y, Stjepanovic, G. | | Deposit date: | 2023-10-12 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation.
Nat Commun, 14, 2023
|
|
7MGE
 
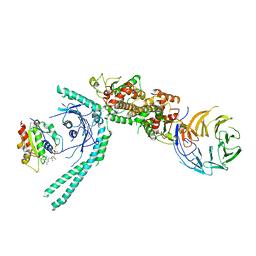 | | Structure of C9orf72:SMCR8:WDR41 in complex with ARF1 | | Descriptor: | ADP-ribosylation factor 1, BERYLLIUM TRIFLUORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Su, M.-Y, Hurley, J.H. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Structural basis for the ARF GAP activity and specificity of the C9orf72 complex.
Nat Commun, 12, 2021
|
|
4R8F
 
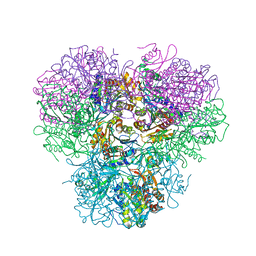 | |
6B9X
 
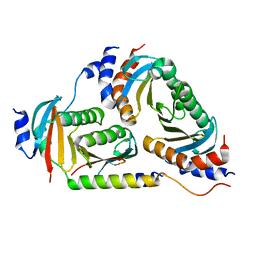 | | Crystal structure of Ragulator | | Descriptor: | Hepatitis B virus x interacting protein, Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, ... | | Authors: | SU, M.-Y, Hurley, J.H. | | Deposit date: | 2017-10-11 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Hybrid Structure of the RagA/C-Ragulator mTORC1 Activation Complex.
Mol. Cell, 68, 2017
|
|
8W4J
 
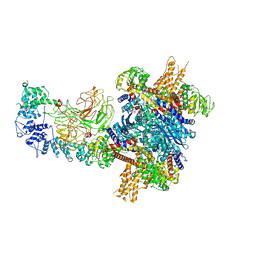 | |
6DCE
 
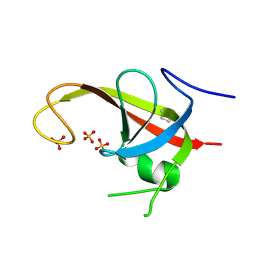 | | X-ray structure of FIP200 claw domain | | Descriptor: | RB1-inducible coiled-coil protein 1, SULFATE ION | | Authors: | Su, M.-Y, Hurley, J.H. | | Deposit date: | 2018-05-05 | | Release date: | 2019-03-06 | | Last modified: | 2019-05-01 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates.
Mol. Cell, 74, 2019
|
|
6GMA
 
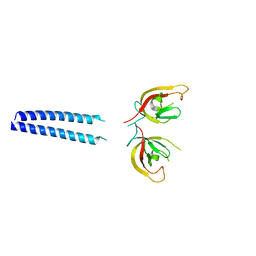 | |
8KHP
 
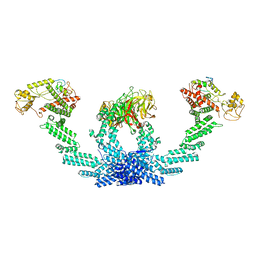 | | CULLIN3-KLHL22-RBX1 E3 ligase | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch-like protein 22 | | Authors: | Su, M.-Y, Su, M.-Y. | | Deposit date: | 2023-08-22 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Cryo-EM structure of the KLHL22 E3 ligase bound to an oligomeric metabolic enzyme.
Structure, 31, 2023
|
|
8KGY
 
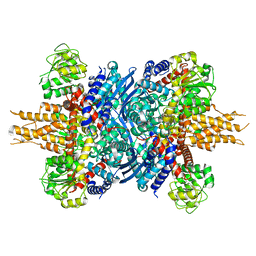 | | Human glutamate dehydrogenase I | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Su, M.-Y. | | Deposit date: | 2023-08-20 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Cryo-EM structure of the KLHL22 E3 ligase bound to an oligomeric metabolic enzyme.
Structure, 31, 2023
|
|
