8BCT
 
 | | X-ray crystal structure of a de novo selected helix-loop-helix heterodimer in a syn arrangement, 26alpha/26beta | | Descriptor: | 26alpha, 26beta, ACETATE ION, ... | | Authors: | Naudin, E.A, Mylemans, B, Smith, A.J, Savery, N.J, Woolfson, D.N. | | Deposit date: | 2022-10-17 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Selection of Heterodimerizing Helical Hairpins for Synthetic Biology.
Acs Synth Biol, 12, 2023
|
|
8BCS
 
 | | X-ray crystal structure of a de novo designed helix-loop-helix homodimer in an anti arrangement, CC-HP1.0 | | Descriptor: | ACETATE ION, CC-HP1.0 | | Authors: | Edgell, C.L, Mylemans, B, Naudin, E.A, Smith, A.J, Savery, N.J, Woolfson, D.N. | | Deposit date: | 2022-10-17 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and Selection of Heterodimerizing Helical Hairpins for Synthetic Biology.
Acs Synth Biol, 12, 2023
|
|
1A9W
 
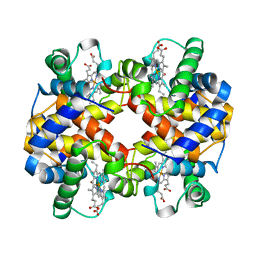 | | HUMAN EMBRYONIC GOWER II CARBONMONOXY HEMOGLOBIN | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), ... | | Authors: | Sutherland-Smith, A.J, Baker, H.M, Hofmann, O.M, Brittain, T, Baker, E.N. | | Deposit date: | 1998-04-11 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a human embryonic haemoglobin: the carbonmonoxy form of gower II (alpha2 epsilon2) haemoglobin at 2.9 A resolution.
J.Mol.Biol., 280, 1998
|
|
4RFL
 
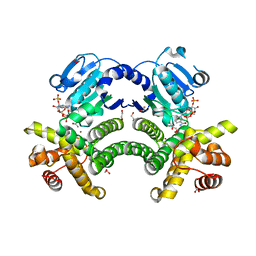 | | Crystal structure of G1PDH with NADPH from Methanocaldococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, Glycerol-1-phosphate dehydrogenase [NAD(P)+], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Carbone, V, Ronimus, R.S, Schofield, L.R, Sutherland-Smith, A.J. | | Deposit date: | 2014-09-26 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Evolution of the Archaeal Lipid Synthesis Enzyme sn-Glycerol-1-phosphate Dehydrogenase.
J.Biol.Chem., 290, 2015
|
|
4RGQ
 
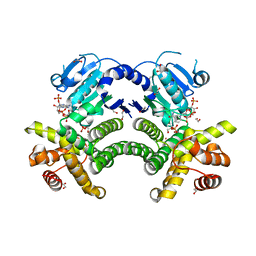 | | Crystal structure of the Methanocaldococcus jannaschii G1PDH with NADPH and DHAP | | Descriptor: | 1,2-ETHANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, Glycerol-1-phosphate dehydrogenase, ... | | Authors: | Carbone, V, Ronimus, R.S, Schofield, L.R, Sutherland-Smith, A.J. | | Deposit date: | 2014-09-30 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure and Evolution of the Archaeal Lipid Synthesis Enzyme sn-Glycerol-1-phosphate Dehydrogenase.
J.Biol.Chem., 290, 2015
|
|
4RGV
 
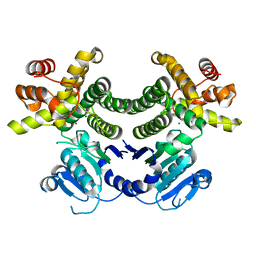 | | Crystal structure of the Methanocaldococcus jannaschii G1PDH | | Descriptor: | Glycerol-1-phosphate dehydrogenase, MAGNESIUM ION, ZINC ION | | Authors: | Carbone, V, Ronimus, R.S, Schofield, L.R, Sutherland-Smith, A.J. | | Deposit date: | 2014-09-30 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and Evolution of the Archaeal Lipid Synthesis Enzyme sn-Glycerol-1-phosphate Dehydrogenase.
J.Biol.Chem., 290, 2015
|
|
7TZI
 
 | | Structure of a pseudomurein peptide ligase type E from Methanothermobacter thermautotrophicus | | Descriptor: | Mur ligase family protein | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.911 Å) | | Cite: | Structural characterisation of methanogen pseudomurein cell wall peptide ligases homologous to bacterial MurE/F murein peptide ligases.
Microbiology (Reading, Engl.), 168, 2022
|
|
7UFP
 
 | | Structure of a pseudomurein peptide ligase type E from Methanothermus fervidus | | Descriptor: | Mur ligase middle domain protein, SULFATE ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | Deposit date: | 2022-03-23 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterisation of methanogen pseudomurein cell wall peptide ligases homologous to bacterial MurE/F murein peptide ligases.
Microbiology (Reading, Engl.), 168, 2022
|
|
3UUM
 
 | |
3UUN
 
 | |
3UUL
 
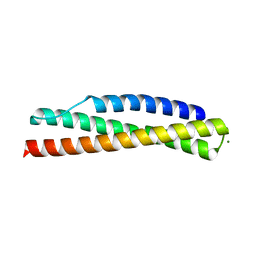 | |
6VR7
 
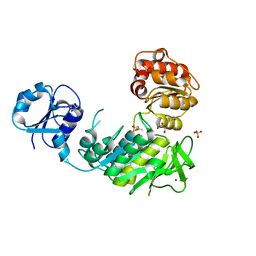 | | Structure of a pseudomurein peptide ligase type C from Methanothermus fervidus | | Descriptor: | ACETOACETIC ACID, GLYCEROL, Mur ligase middle domain protein, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Archaeal pseudomurein and bacterial murein cell wall biosynthesis share a common evolutionary ancestry
FEMS Microbes, 2, 2021
|
|
6VR8
 
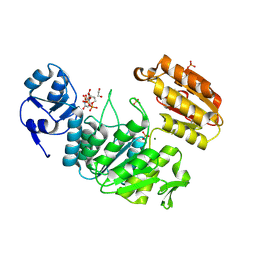 | | Structure of a pseudomurein peptide ligase type E from Methanothermus fervidus | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Mur ligase middle domain protein, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | Deposit date: | 2020-02-06 | | Release date: | 2021-08-11 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterisation of methanogen pseudomurein cell wall peptide ligases homologous to bacterial MurE/F murein peptide ligases.
Microbiology (Reading, Engl.), 168, 2022
|
|
6P8C
 
 | | 2,5-diamino-6-(ribosylamino)-4(3H)-pyrimidinone 5'-phosphate reductase (MthRED) from Methanothermobacter thermautotrophicus | | Descriptor: | 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5'-phosphate reductase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Carbone, V, Schofield, L.R, Hannus, I, Sutherland-Smith, A.J, Ronimus, R.S. | | Deposit date: | 2019-06-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The Crystal Structure of 2,5-diamino-6-(ribosylamino)-4(3H)-pyrimidinone 5'-phosphate reductase (MthRED) from Methanothermobacter thermautotrophicus
To Be Published
|
|
6PNL
 
 | | Structure of Epimerase Mth375 from the thermophilic pseudomurein-containing methanogen Methanothermobacter thermautotrophicus | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of a UDP-GALE 4-epimerase (Mth375) from the thermophilic pseudomurein-containing methanogen Methanothermobacter thermautotrophicus
To Be Published
|
|
6PMH
 
 | | Structure of Epimerase Mth375 from the thermophilic pseudomurein-containing methanogen Methanothermobacter thermautotrophicus | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S. | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a UDP-GALE 4-epimerase (Mth375) from the thermophilic pseudomurein-containing methanogen Methanothermobacter thermautotrophicus
To Be Published
|
|
3HK1
 
 | | Identification and Characterization of a Small Molecule Inhibitor of Fatty Acid Binding Proteins | | Descriptor: | 4-{[2-(methoxycarbonyl)-5-(2-thienyl)-3-thienyl]amino}-4-oxo-2-butenoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Hertzel, A.V, Hellberg, K, Reynolds, J.M, Kruse, A.C, Juhlmann, B.E, Smith, A.J, Sanders, M.A, Ohlendorf, D.H, Suttles, J, Bernlohr, D.A. | | Deposit date: | 2009-05-22 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and characterization of a small molecule inhibitor of Fatty Acid binding proteins.
J.Med.Chem., 52, 2009
|
|
3HOP
 
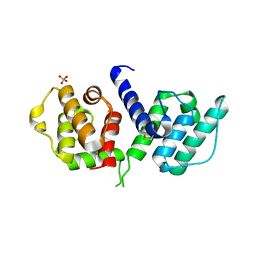 | | Structure of the actin-binding domain of human filamin A | | Descriptor: | Filamin-A, PHOSPHATE ION | | Authors: | Clark, A.R, Sawyer, G.M, Robertson, S.P, Sutherland-Smith, A.J. | | Deposit date: | 2009-06-03 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Skeletal dysplasias due to filamin A mutations result from a gain-of-function mechanism distinct from allelic neurological disorders
Hum.Mol.Genet., 18, 2009
|
|
3HOR
 
 | | Structure of the actin-binding domain of human filamin A (reduced) | | Descriptor: | Filamin-A, PHOSPHATE ION | | Authors: | Clark, A.R, Sawyer, G.M, Robertson, S.P, Sutherland-Smith, A.J. | | Deposit date: | 2009-06-03 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Skeletal dysplasias due to filamin A mutations result from a gain-of-function mechanism distinct from allelic neurological disorders
Hum.Mol.Genet., 18, 2009
|
|
3HOC
 
 | | Structure of the actin-binding domain of human filamin A mutant E254K | | Descriptor: | Filamin-A, PHOSPHATE ION | | Authors: | Clark, A.R, Sawyer, G.M, Robertson, S.P, Sutherland-Smith, A.J. | | Deposit date: | 2009-06-02 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Skeletal dysplasias due to filamin A mutations result from a gain-of-function mechanism distinct from allelic neurological disorders
Hum.Mol.Genet., 18, 2009
|
|
4FIO
 
 | | Crystal Structure of Methenyltetrahydromethanopterin Cyclohydrolase from Methanobrevibacter ruminantium | | Descriptor: | ETHYL ACETATE, ISOPROPYL ALCOHOL, Methenyltetrahydromethanopterin cyclohydrolase | | Authors: | Carbone, V, Schofield, L.R, Beattie, A.K, Sutherland-Smith, A.J, Ronimus, R.S. | | Deposit date: | 2012-06-10 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The crystal structure of methenyltetrahydromethanopterin cyclohydrolase from Methanobrevibacter ruminantium.
Proteins, 81, 2013
|
|
6DNT
 
 | | UDP-N-acetylglucosamine 4-epimerase from Methanobrevibacter ruminantium M1 in complex with UDP-N-acetylmuramic acid | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase, ... | | Authors: | Carbone, V, Schofield, L.R, Sang, C, Sutherland-Smith, A.J, Ronimus, R.S. | | Deposit date: | 2018-06-07 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural determination of archaeal UDP-N-acetylglucosamine 4-epimerase from Methanobrevibacter ruminantium M1 in complex with the bacterial cell wall intermediate UDP-N-acetylmuramic acid.
Proteins, 86, 2018
|
|
1DXX
 
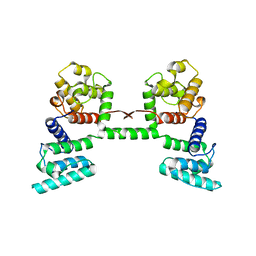 | |
4GMH
 
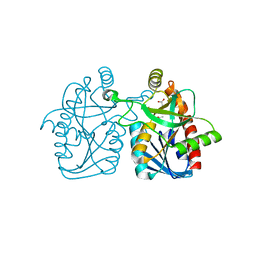 | | Crystal structure of staphylococcus aureus 5'-methylthioadenosine/s-adenosylhomocysteine nucleosidase | | Descriptor: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ACETATE ION | | Authors: | Brown, R.L, Anderson, B.F, Norris, G.E, Tyler, P.C, Evans, G.B, Sutherland-Smith, A.J. | | Deposit date: | 2012-08-15 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of staphylococcus aureus 5'-methylthioadenosine/s-adenosylhomocysteine nucleosidase
To be Published
|
|
5CK7
 
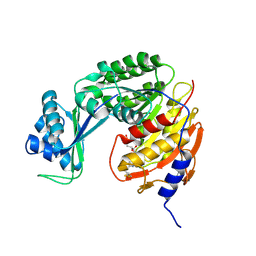 | |
