4QPZ
 
 | | Crystal structure of the formolase FLS_v2 in space group P 21 | | Descriptor: | Formolase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Shen, B.W, Siegel, J.B, Stoddard, B.L, Baker, D. | | Deposit date: | 2014-06-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Computational protein design enables a novel one-carbon assimilation pathway.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4QQ8
 
 | | Crystal structure of the formolase FLS in space group P 43 21 2 | | Descriptor: | 1,2-ETHANEDIOL, Formolase, MAGNESIUM ION, ... | | Authors: | Shen, B.W, Siegel, J.B, Stoddard, B.L. | | Deposit date: | 2014-06-26 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Computational protein design enables a novel one-carbon assimilation pathway.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8FHB
 
 | |
8FHC
 
 | | Protein 41 with aldehyde deformylating oxidase activity from Gamma proteobacterium | | Descriptor: | BROMIDE ION, CHOLIC ACID, FE (III) ION, ... | | Authors: | Arenas, R, Wilson, D.K, Mak, W.S, Siegel, J.B. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Protein 41 with aldehyde deformylating oxidase activity from Gamma proteobacterium
To Be Published
|
|
6D9F
 
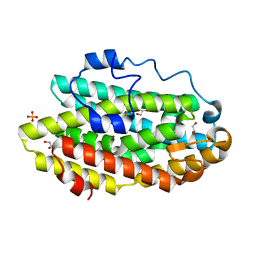 | | Protein 60 with aldehyde deformylating oxidase activity from Kitasatospora setae | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Putative VlmB homolog, ... | | Authors: | Arenas, R, Wilson, D.K, Mak, W.S, Siegel, J.B. | | Deposit date: | 2018-04-28 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery, Design, and Structural Characterization of Alkane-Producing Enzymes across the Ferritin-like Superfamily.
Biochemistry, 59, 2020
|
|
5UX1
 
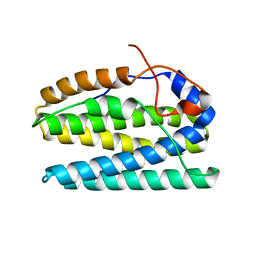 | |
5UXI
 
 | |
5UX2
 
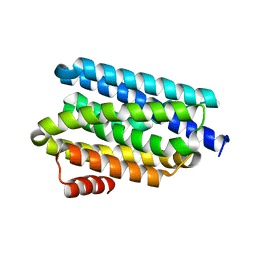 | |
5UXG
 
 | |
5V4T
 
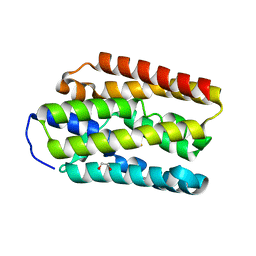 | |
3I1C
 
 | |
3U0S
 
 | | Crystal Structure of an Enzyme Redesigned Through Multiplayer Online Gaming: CE6 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Diisopropyl-fluorophosphatase, GLYCEROL, ... | | Authors: | Bale, J.B, Shen, B.W, Stoddard, B.L. | | Deposit date: | 2011-09-29 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Increased Diels-Alderase activity through backbone remodeling guided by Foldit players.
Nat.Biotechnol., 30, 2012
|
|
6VPT
 
 | |
7T1J
 
 | | Crystal structure of RUBISCO from Rhodospirillaceae bacterium BRH_c57 | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Pereira, J.H, Liu, A.K, Shih, P.M, Adams, P.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural plasticity enables evolution and innovation of RuBisCO assemblies.
Sci Adv, 8, 2022
|
|
7T1C
 
 | |
4O5T
 
 | | Crystal structure of Diels-Alderase CE20 in complex with a product analog | | Descriptor: | 4-{[2-(phosphonooxy)ethyl]carbamoyl}benzyl [(1R,6S)-6-(dimethylcarbamoyl)cyclohex-2-en-1-yl]carbamate, Diisopropyl-fluorophosphatase | | Authors: | Beck, T, Preiswerk, N, Mayer, C, Hilvert, D. | | Deposit date: | 2013-12-20 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Impact of scaffold rigidity on the design and evolution of an artificial Diels-Alderase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4O5S
 
 | |
5UWZ
 
 | |
4F5J
 
 | |
4F5F
 
 | |
4F5M
 
 | |
4F5I
 
 | |
4F5H
 
 | |
4F5L
 
 | |
4F5K
 
 | |
