8J9R
 
 | |
6IVH
 
 | |
5XNS
 
 | |
5XG3
 
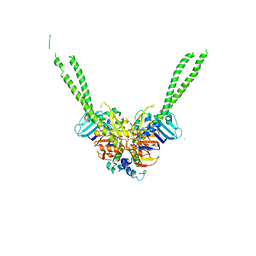 | | Crystal structure of the ATPgS-engaged Smc head domain with an extended coiled coil bound to the C-terminal domain of ScpA derived from Bacillus subtilis | | Descriptor: | COBALT (II) ION, Chromosome partition protein Smc, MAGNESIUM ION, ... | | Authors: | Shin, H.-C, Lee, H, Oh, B.-H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization
Mol. Cell, 67, 2017
|
|
8J9Q
 
 | | Crystal structure of UBR box of UBR4 apo | | Descriptor: | E3 ubiquitin-protein ligase UBR4, ZINC ION | | Authors: | Jeong, D.-E, KIm, S.-J, Shin, H.-C. | | Deposit date: | 2023-05-04 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Insights into the recognition mechanism in the UBR box of UBR4 for its specific substrates.
Commun Biol, 6, 2023
|
|
6IM5
 
 | | YAP-binding domain of human TEAD1 | | Descriptor: | PHOSPHATE ION, Transcriptional enhancer factor TEF-1 | | Authors: | Mo, Y, Lee, H.S, Kim, S.J, Ku, B. | | Deposit date: | 2018-10-22 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Crystal Structure of the YAP-binding Domain of Human TEAD1
Bull.Korean Chem.Soc., 40, 2019
|
|
6JUV
 
 | |
5GOT
 
 | | Crystal structure of SP-PTP, low molecular weight protein tyrosine phosphatase from Streptococcus pyogenes | | Descriptor: | CHLORIDE ION, Low molecular weight phosphotyrosine phosphatase family protein | | Authors: | Ku, B, Keum, C.W, KIim, S.J. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal structure of SP-PTP, a low molecular weight protein tyrosine phosphatase from Streptococcus pyogenes
Biochem.Biophys.Res.Commun., 478, 2016
|
|
2TNF
 
 | | 1.4 A RESOLUTION STRUCTURE OF MOUSE TUMOR NECROSIS FACTOR, TOWARDS MODULATION OF ITS SELECTIVITY AND TRIMERISATION | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ISOPROPYL ALCOHOL, PROTEIN (TUMOR NECROSIS FACTOR ALPHA) | | Authors: | Baeyens, K.J, De Bondt, H.L, Raeymaekers, A, Fiers, W, De Ranter, C.J. | | Deposit date: | 1998-10-12 | | Release date: | 1999-10-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of mouse tumour-necrosis factor at 1.4 A resolution: towards modulation of its selectivity and trimerization.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
