1U0L
 
 | | Crystal structure of YjeQ from Thermotoga maritima | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Probable GTPase engC, ZINC ION | | Authors: | Shin, D.H, Lou, Y, Jaru, J, Kim, R, Yokota, H, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-07-13 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of YjeQ from Thermotoga maritima contains a circularly permuted GTPase domain
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
2HY5
 
 | | Crystal structure of DsrEFH | | Descriptor: | DsrH, Intracellular sulfur oxidation protein dsrF, Putative sulfurtransferase dsrE | | Authors: | Shin, D.H, Schulte, A, Dahl, C, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of DsrEFH
To be Published
|
|
2I1O
 
 | | Crystal Structure of a Nicotinate Phosphoribosyltransferase from Thermoplasma acidophilum | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Shin, D.H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-08-14 | | Release date: | 2006-08-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a zinc ion bound Nicotinate Phosphoribosyltransferase from Thermoplasma acidophilum
To be Published
|
|
2I14
 
 | |
8HHD
 
 | | Crystal structure of PaMurU | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nucleotidyl transferase, SULFATE ION | | Authors: | Shin, D.H, Jo, S.R, Kim, M.S. | | Deposit date: | 2022-11-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of PAMurU
To Be Published
|
|
1MZL
 
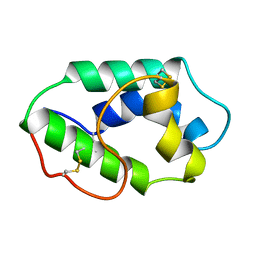 | | MAIZE NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | MAIZE NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Shin, D.H, Lee, J.Y, Hwang, K.Y, Kim, K.K, Suh, S.W. | | Deposit date: | 1995-01-26 | | Release date: | 1996-08-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structure of the non-specific lipid-transfer protein from maize seedlings.
Structure, 3, 1995
|
|
6IZB
 
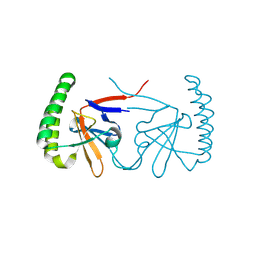 | | Flexible loop truncated human TCTP | | Descriptor: | Translationally-controlled tumor protein | | Authors: | Shin, D.H, Kim, M.S. | | Deposit date: | 2018-12-19 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Flexible loop truncated human TCTP
To Be Published
|
|
1YTK
 
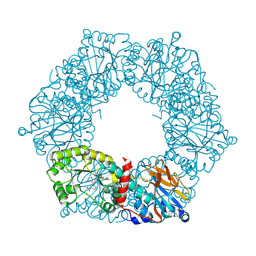 | |
1YTD
 
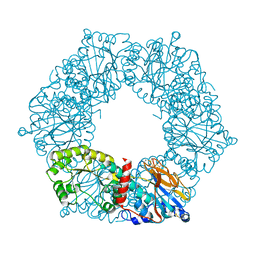 | |
1YTE
 
 | |
6IZE
 
 | | Dimeric human TCTP | | Descriptor: | Translationally-controlled tumor protein | | Authors: | Shin, D.H, Kim, M.S. | | Deposit date: | 2018-12-19 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Dimeric human TCTP
To Be Published
|
|
2BA2
 
 | | Crystal structure of the DUF16 domain of MPN010 from Mycoplasma pneumoniae | | Descriptor: | Hypothetical UPF0134 protein MPN010 | | Authors: | Shin, D.H, Kim, J.-S, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-10-13 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the DUF16 domain of MPN010 from Mycoplasma pneumoniae.
Protein Sci., 15, 2006
|
|
2HYB
 
 | | Crystal Structure of Hexameric DsrEFH | | Descriptor: | DsrH, Intracellular sulfur oxidation protein dsrF, Putative sulfurtransferase dsrE | | Authors: | Shin, D.H, Connie, H, Schulte, A, Dahl, C, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-08-04 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hexameric DsrEFH
To be Published
|
|
2I15
 
 | |
1NF2
 
 | |
1L2F
 
 | | Crystal structure of NusA from Thermotoga maritima: a structure-based role of the N-terminal domain | | Descriptor: | N utilization substance protein A | | Authors: | Shin, D.H, Nguyen, H.H, Jancarik, J, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-02-20 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of NusA from Thermotoga maritima and functional implication of the N-terminal domain.
Biochemistry, 42, 2003
|
|
1LFP
 
 | | Crystal Structure of a Conserved Hypothetical Protein Aq1575 from Aquifex Aeolicus | | Descriptor: | Hypothetical protein AQ_1575 | | Authors: | Shin, D.H, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-04-11 | | Release date: | 2002-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of conserved hypothetical protein Aq1575 from Aquifex aeolicus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1NYE
 
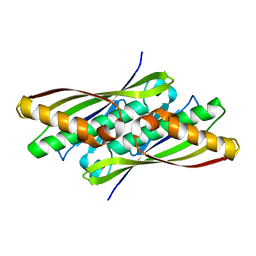 | | Crystal structure of OsmC from E. coli | | Descriptor: | Osmotically inducible protein C | | Authors: | Shin, D.H, Choi, I.-G, Busso, D, Jancarik, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-02-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of OsmC from Escherichia coli: a salt-shock-induced protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ECY
 
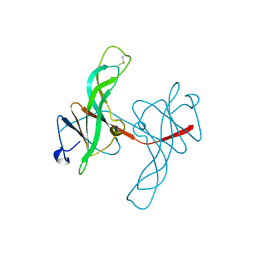 | | PROTEASE INHIBITOR ECOTIN | | Descriptor: | ECOTIN, alpha-D-glucopyranose, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | Authors: | Shin, D.H, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure analyses of uncomplexed ecotin in two crystal forms: implications for its function and stability.
Protein Sci., 5, 1996
|
|
1ECZ
 
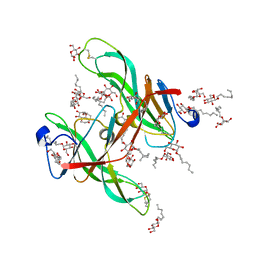 | | PROTEASE INHIBITOR ECOTIN | | Descriptor: | ECOTIN, octyl beta-D-glucopyranoside | | Authors: | Shin, D.H, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure analyses of uncomplexed ecotin in two crystal forms: implications for its function and stability.
Protein Sci., 5, 1996
|
|
1S7C
 
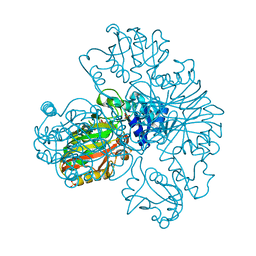 | | Crystal structure of MES buffer bound form of glyceraldehyde 3-phosphate dehydrogenase from Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glyceraldehyde 3-phosphate dehydrogenase A, SULFATE ION | | Authors: | Shin, D.H, Thor, J, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-29 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of MES buffer bound form of glyceraldehyde 3-phosphate dehydrogenase from Escherichia coli
To be Published
|
|
1T71
 
 | | Crystal structure of a novel phosphatase Mycoplasma pneumoniaefrom | | Descriptor: | FE (III) ION, phosphatase | | Authors: | Shin, D.H, Jancarik, J, Kim, R, Yokota, H, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-07 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a novel phosphatase from Mycoplasma pneumoniae
To be Published
|
|
1JX7
 
 | | Crystal structure of ychN protein from E.coli | | Descriptor: | HYPOTHETICAL PROTEIN YCHN | | Authors: | Shin, D.H, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2001-09-05 | | Release date: | 2002-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a conserved hypothetical protein from Escherichia coli
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
1S7D
 
 | |
1S12
 
 | | Crystal structure of TM1457 | | Descriptor: | ACETATE ION, hypothetical protein TM1457 | | Authors: | Shin, D.H, Lou, Y, Jancarik, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-05 | | Release date: | 2004-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of TM1457 from Thermotoga maritima.
J.Struct.Biol., 152, 2005
|
|
