5HNY
 
 | | Structural basis of backwards motion in kinesin-14: plus-end directed nKn669 in the AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2016-11-02 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
5HNZ
 
 | | Structural basis of backwards motion in kinesin-14: plus-end directed nKn669 in the nucleotide-free state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
5HNX
 
 | | Structural basis of backwards motion in kinesin-14: minus-end directed nKn664 in the nucleotide-free state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
5HNW
 
 | | Structural basis of backwards motion in kinesin-14: minus-end directed nKn664 in the AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2018-07-25 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
8XI6
 
 | | SARS-CoV-2 Omicron BQ.1.1 Variant Spike Protein Complexed with MO11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ishimaru, H, Nishimura, M, Shigematsu, H, Marini, M.I, Hasegawa, N, Takamiya, R, Iwata, S, Mori, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Epitopes of an antibody that neutralizes a wide range of SARS-CoV-2 variants in a conserved subdomain 1 of the spike protein.
J.Virol., 2024
|
|
1V7M
 
 | | Human Thrombopoietin Functional Domain Complexed To Neutralizing Antibody TN1 Fab | | Descriptor: | Monoclonal TN1 Fab Heavy Chain, Monoclonal TN1 Fab Light Chain, Thrombopoietin | | Authors: | Feese, M.D, Tamada, T, Kato, Y, Maeda, Y, Hirose, M, Matsukura, Y, Shigematsu, H, Kato, T, Miyazaki, H, Kuroki, R. | | Deposit date: | 2003-12-18 | | Release date: | 2004-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of the receptor-binding domain of human thrombopoietin determined by complexation with a neutralizing antibody fragment
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1V7N
 
 | | Human Thrombopoietin Functional Domain Complexed To Neutralizing Antibody TN1 Fab | | Descriptor: | Monoclonal TN1 Fab Heavy Chain, Monoclonal TN1 Fab Light Chain, Thrombopoietin | | Authors: | Feese, M.D, Tamada, T, Kato, Y, Maeda, Y, Hirose, M, Matsukura, Y, Shigematsu, H, Kato, T, Miyazaki, H, Kuroki, R. | | Deposit date: | 2003-12-18 | | Release date: | 2004-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the receptor-binding domain of human thrombopoietin determined by complexation with a neutralizing antibody fragment
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8WUC
 
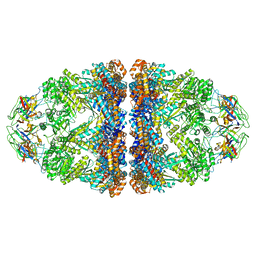 | | Cryo-EM structure of H. thermoluteolus GroEL-GroES2 football complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 2024
|
|
8WUW
 
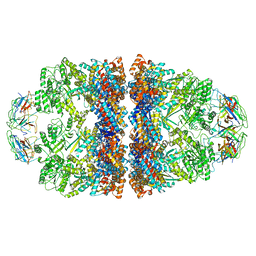 | | Cryo-EM structure of H. thermophilus GroEL-GroES2 asymmetric football complex | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, MAGNESIUM ION, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-21 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 2024
|
|
8WU4
 
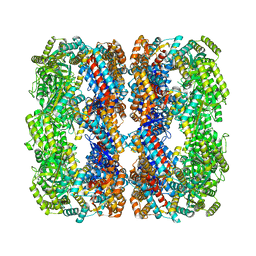 | | Cryo-EM structure of native H. thermoluteolus TH-1 GroEL | | Descriptor: | Chaperonin GroEL | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 2024
|
|
8WUX
 
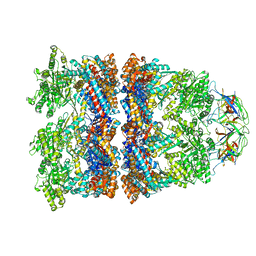 | | Cryo-EM structure of H. thermophilus GroEL-GroES bullet complex | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, MAGNESIUM ION, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-21 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 2024
|
|
8H3M
 
 | | Conformation 1 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy chain, Spike glycoprotein | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
8H3N
 
 | | Conformation 2 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy-chain, MO1 light chain, ... | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
5GSY
 
 | | Kinesin-8 motor, KIF19A, in the nucleotide-free state complexed with GDP-taxol microtubule | | Descriptor: | Kinesin-like protein KIF19 | | Authors: | Morikawa, M, Nitta, R, Yajima, H, Shigematsu, H, Kikkawa, M, Hirokawa, N. | | Deposit date: | 2016-08-17 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A
Elife, 5, 2016
|
|
8IPB
 
 | | Wheat 80S ribosome pausing on AUG-Stop with cycloheximide | | Descriptor: | 18S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, 40S ribosomal protein eL8, ... | | Authors: | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 2024
|
|
8IP9
 
 | | Wheat 40S ribosome in complex with a tRNAi | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S23, 40S ribosomal protein eS1, ... | | Authors: | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 2024
|
|
8IP8
 
 | | Wheat 80S ribosome stalled on AUG-Stop boron dependently | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein eL8, 40S ribosomal protein eS1, ... | | Authors: | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 2024
|
|
8IPA
 
 | | Wheat 80S ribosome stalled on AUG-Stop boron dependently with cycloheximide | | Descriptor: | 18S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, 40S ribosomal protein eL8, ... | | Authors: | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 2024
|
|
7XMC
 
 | | Cryo-EM structure of Cytochrome bo3 from Escherichia coli, apo structure with DMSO | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Nishida, Y, Shigematsu, H, Iwamoto, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7XMD
 
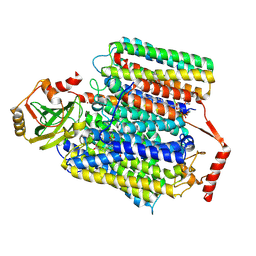 | | Cryo-EM structure of Cytochrome bo3 from Escherichia coli, the structure complexed with an allosteric inhibitor N4 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Nishida, Y, Shigematsu, H, Iwamoto, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
5G2X
 
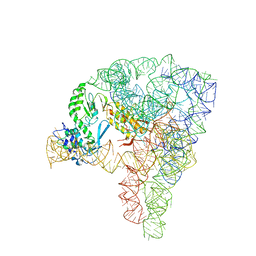 | | Structure a of Group II Intron Complexed with its Reverse Transcriptase | | Descriptor: | 5'-R(*CP*AP*CP*AP*UP*CP*CP*AP*UP*AP*AP*CP)-3', GROUP II INTRON, GROUP II INTRON-ENCODED PROTEIN LTRA | | Authors: | Qu, G, Kaushal, P.S, Wang, J, Shigematsu, H, Piazza, C.L, Agrawal, R.K, Belfort, M, Wang, H.W. | | Deposit date: | 2016-04-16 | | Release date: | 2016-05-11 | | Last modified: | 2016-06-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a Group II Intron in Complex with its Reverse Transcriptase.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5G2Y
 
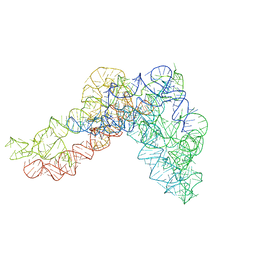 | | Structure a of Group II Intron Complexed with its Reverse Transcriptase | | Descriptor: | GROUP II INTRON | | Authors: | Qu, G, Kaushal, P.S, Wang, J, Shigematsu, H, Piazza, C.L, Agrawal, R.K, Belfort, M, Wang, H.W. | | Deposit date: | 2016-04-16 | | Release date: | 2016-05-04 | | Last modified: | 2016-06-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of a Group II Intron in Complex with its Reverse Transcriptase.
Nat.Struct.Mol.Biol., 23, 2016
|
|
7D7M
 
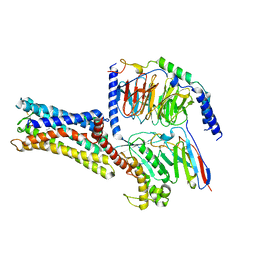 | | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Nojima, S, Fujita, Y, Kimura, T.K, Nomura, N, Suno, R, Morimoto, K, Yamamoto, M, Noda, T, Iwata, S, Shigematsu, H, Kobayashi, T. | | Deposit date: | 2020-10-05 | | Release date: | 2020-11-18 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein.
Structure, 29, 2021
|
|
6IP6
 
 | | Cryo-EM structure of the CMV-stalled human 80S ribosome with HCV IRES (Structure iii) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | Deposit date: | 2018-11-02 | | Release date: | 2019-05-29 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
6IP8
 
 | | Cryo-EM structure of the HCV IRES dependently initiated CMV-stalled 80S ribosome (Structure iv) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | Deposit date: | 2018-11-02 | | Release date: | 2019-05-29 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
