6M6B
 
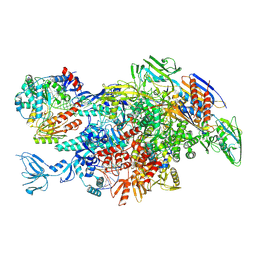 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase and ATP-gamma-S | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6M6A
 
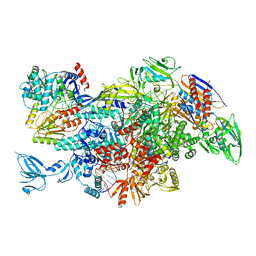 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6M6C
 
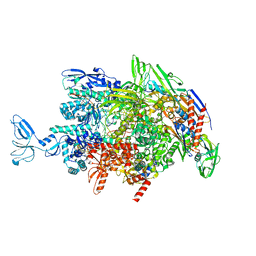 | | CryoEM structure of Thermus thermophilus RNA polymerase elongation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
5LUS
 
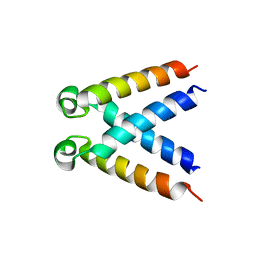 | | Structures of DHBN domain of Pelecanus crispus BLM helicase | | Descriptor: | BLM helicase | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.433 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
5LUP
 
 | | Structures of DHBN domain of human BLM helicase | | Descriptor: | BLM protein, PHOSPHATE ION, POTASSIUM ION | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
5LUT
 
 | | Structures of DHBN domain of Gallus gallus BLM helicase | | Descriptor: | BLM helicase, PHOSPHATE ION | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
5GNG
 
 | |
3IKK
 
 | | Crystal structure analysis of msp domain | | Descriptor: | Vesicle-associated membrane protein-associated protein B/C | | Authors: | Shi, J, Lua, S, Song, J. | | Deposit date: | 2009-08-06 | | Release date: | 2010-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Elimination of the native structure and solubility of the hVAPB MSP domain by the Pro56Ser mutation that causes amyotrophic lateral sclerosis.
Biochemistry, 49, 2010
|
|
5H3B
 
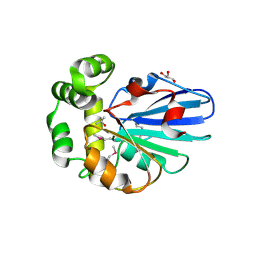 | |
6J31
 
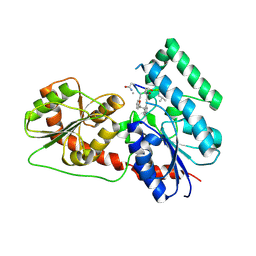 | | Crystal Structure Analysis of the Glycotransferase of kitacinnamycin | | Descriptor: | (2E,2'E)-3,3'-(1,2-phenylene)di(prop-2-enoic acid), DBB-DSG-VAL-MEA-VAL-GLY-GLY-DVA-DLE, kcn28 | | Authors: | Shi, J, Liu, C.L, Zhang, B, Guo, W.J, Zhu, J.P, Xu, X, Xu, Q, Jiao, R.H, Tan, R.X, Ge, H.M. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Genome mining and biosynthesis of kitacinnamycins as a STING activator.
Chem Sci, 10, 2019
|
|
6J32
 
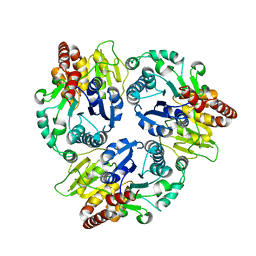 | | Crystal Structure Analysis of the Glycotransferase of kitacinnamycin | | Descriptor: | Kcn28 | | Authors: | Shi, J, Liu, C.L, Zhang, B, Guo, W.J, Zhu, J.P, Xu, X, Xu, Q, Jiao, R.H, Tan, R.X, Ge, H.M. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Genome mining and biosynthesis of kitacinnamycins as a STING activator.
Chem Sci, 10, 2019
|
|
6K4Y
 
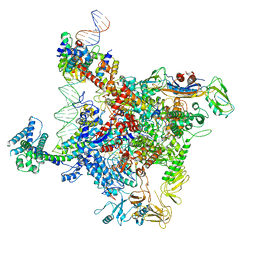 | | CryoEM structure of sigma appropriation complex | | Descriptor: | 10 kDa anti-sigma factor, DNA (60-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis of sigma appropriation.
Nucleic Acids Res., 47, 2019
|
|
7D7D
 
 | | CryoEM structure of gp45-dependent transcription activation complex | | Descriptor: | DNA (nontemplate strand), DNA (template strand), DNA polymerase clamp, ... | | Authors: | Shi, J, Wen, A, Jin, S, Feng, Y. | | Deposit date: | 2020-10-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Transcription activation by a sliding clamp.
Nat Commun, 12, 2021
|
|
7D7C
 
 | | CryoEM structure of gp55-dependent RNA polymerase-promoter open complex | | Descriptor: | DNA (nontemplate strand), DNA (template strand), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shi, J, Wen, A, Jin, S, Feng, Y. | | Deposit date: | 2020-10-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Transcription activation by a sliding clamp.
Nat Commun, 12, 2021
|
|
5MK5
 
 | | Structures of DHBN domain of human BLM helicase | | Descriptor: | Bloom syndrome protein, IODIDE ION, POTASSIUM ION | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-12-02 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
2VLM
 
 | | The Structural Dynamics and Energetics of an Immunodominant T-cell Receptor are Programmed by its Vbeta Domain | | Descriptor: | JM22 TCR ALPHA CHAIN, JM22 TCR BETA CHAIN | | Authors: | Ishizuka, J, Stewart-Jones, G, Van der Merwe, A, Bell, J, McMichael, A, Jones, Y. | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Structural Dynamics and Energetics of an Immunodominant T-Cell Receptor are Programmed by its Vbeta Domain
Immunity, 28, 2008
|
|
5XW4
 
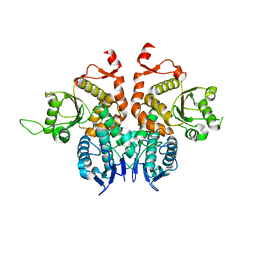 | |
5XW5
 
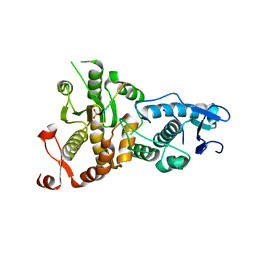 | |
3DQS
 
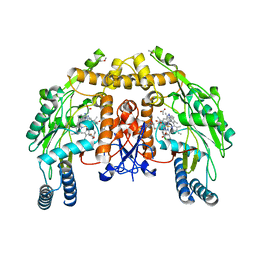 | | Structure of endothelial NOS heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2008-07-09 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
3DQR
 
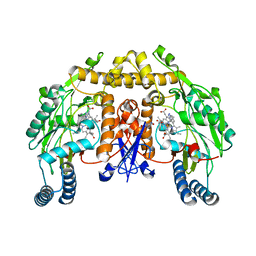 | | Structure of neuronal NOS D597N/M336V mutant heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-aminopyridin-2"-yl)methyl]pyrrolidin-3'-yl}ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-aminopyridin-2-yl)methyl]pyrrolidin-3-yl}ethane-1,2-diamine, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2008-07-09 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
3DQT
 
 | | Structure of endothelial NOS heme domain in complex with a inhibitor (+-)-N1-{trans-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2008-07-09 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
6LXG
 
 | | NMR solution structure of regulatory ACT domain of the Mycobacterium tuberculosis Rel protein | | Descriptor: | GTP pyrophosphokinase | | Authors: | Shin, J, Singal, B, Manimekalai, M.S.S, Gruber, G. | | Deposit date: | 2020-02-11 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Atomic structure of, and valine binding to the regulatory ACT domain of the Mycobacterium tuberculosis Rel protein.
Febs J., 288, 2021
|
|
8WFB
 
 | | Cryo-EM structure of the dPspCas13b-ADAR2-crRNA-target RNA complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, ZINC ION, crRNA, ... | | Authors: | Ishikawa, J, Kato, K, Yamashita, K, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2023-09-19 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insights into RNA-guided RNA editing by the Cas13b-ADAR2 complex.
Nat.Struct.Mol.Biol., 2025
|
|
8WF8
 
 | | Cryo-EM structure of the PspCas13b-crRNA complex | | Descriptor: | MAGNESIUM ION, PspCas13b, crRNA | | Authors: | Ishikawa, J, Kato, K, Yamashita, K, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2023-09-19 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural insights into RNA-guided RNA editing by the Cas13b-ADAR2 complex.
Nat.Struct.Mol.Biol., 2025
|
|
8WF9
 
 | | Cryo-EM structure of the PspCas13b-crRNA-target RNA complex (State 1) | | Descriptor: | MAGNESIUM ION, PspCas13b, crRNA, ... | | Authors: | Ishikawa, J, Kato, K, Yamashita, K, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2023-09-19 | | Release date: | 2025-03-19 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural insights into RNA-guided RNA editing by the Cas13b-ADAR2 complex.
Nat.Struct.Mol.Biol., 2025
|
|
