3ZC8
 
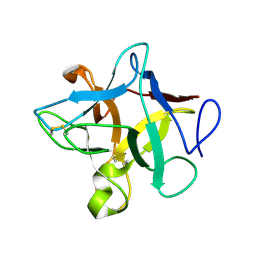 | | Crystal Structure of Murraya koenigii Miraculin-Like Protein at 2.2 A resolution at pH 7.0 | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Selvakumar, P, Sharma, N, Tomar, P.P.S, Kumar, P, Sharma, A.K. | | Deposit date: | 2012-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Insights Into the Aggregation Behavior of Murraya Koenigii Miraculin-Like Protein Below Ph 7.5.
Proteins, 82, 2014
|
|
3ZC9
 
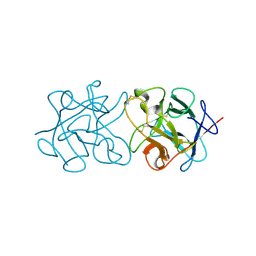 | | Crystal Structure of Murraya koenigii Miraculin-Like Protein at 2.2 A resolution at pH 4.6 | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Selvakumar, P, Sharma, N, Tomar, P.P.S, Kumar, P, Sharma, A.K. | | Deposit date: | 2012-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Insights Into the Aggregation Behavior of Murraya Koenigii Miraculin-Like Protein Below Ph 7.5.
Proteins, 82, 2014
|
|
8DFL
 
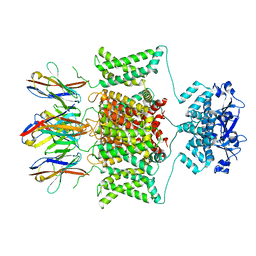 | |
7LVT
 
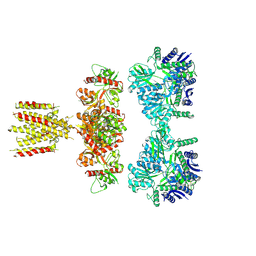 | | Structure of full-length GluK1 with L-Glu | | Descriptor: | Isoform Glur5-2 of Glutamate receptor ionotropic, kainate 1 | | Authors: | Meyerson, J.R, Selvakumar, P. | | Deposit date: | 2021-02-26 | | Release date: | 2021-11-03 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural and compositional diversity in the kainate receptor family.
Cell Rep, 37, 2021
|
|
7SSZ
 
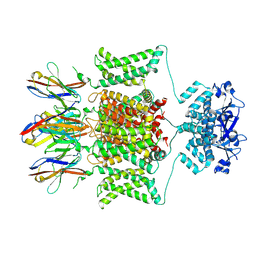 | | Structure of human Kv1.3 with A0194009G09 nanobodies | | Descriptor: | Nanobody A0194009G09, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3,Green fluorescent protein fusion | | Authors: | Meyerson, J.R, Selvakumar, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
7SSV
 
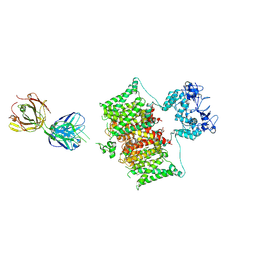 | | Structure of human Kv1.3 with Fab-ShK fusion | | Descriptor: | Fab-ShK fusion, heavy chain, light chain, ... | | Authors: | Meyerson, J.R, Selvakumar, P, Smider, V, Huang, R. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
7SSY
 
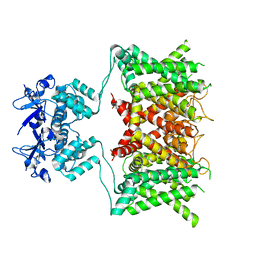 | | Structure of human Kv1.3 (alternate conformation) | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3,Green fluorescent protein fusion | | Authors: | Meyerson, J.R, Selvakumar, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
7SSX
 
 | | Structure of human Kv1.3 | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, Green fluorescent protein fusion | | Authors: | Meyerson, J.R, Selvakumar, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
3IIR
 
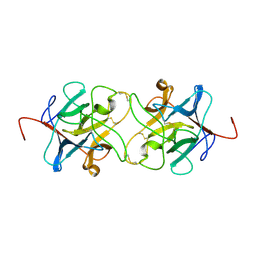 | | Crystal Structure of Miraculin like protein from seeds of Murraya koenigii | | Descriptor: | Trypsin inhibitor | | Authors: | Gahloth, D, Selvakumar, P, Shee, C, Kumar, P, Sharma, A.K. | | Deposit date: | 2009-08-03 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cloning, sequence analysis and crystal structure determination of a miraculin-like protein from Murraya koenigii
Arch.Biochem.Biophys., 494, 2010
|
|
5AFS
 
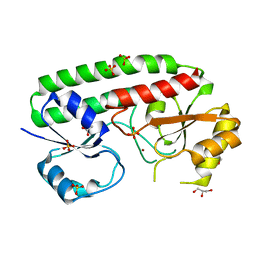 | | structure of Zn-bound periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, GLYCEROL, PERIPLASMIC SOLUTE BINDING PROTEIN, ... | | Authors: | Sharma, N, Selvakumar, P, Kumar, P, Sharma, A.K. | | Deposit date: | 2015-01-23 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure analysis in Zn(2+)-bound state and biophysical characterization of CLas-ZnuA2.
Biochim. Biophys. Acta, 1864, 2016
|
|
4UDN
 
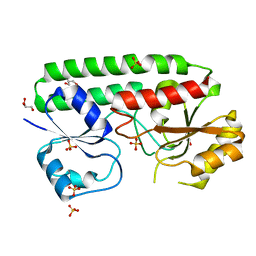 | | structure of metal-free periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, GLYCEROL, PERIPLASMIC SOLUTE BINDING PROTEIN, ... | | Authors: | Sharma, N, Selvakumar, P, Kumar, P, Sharma, A.K. | | Deposit date: | 2014-12-10 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of a Periplasmic Solute Binding Protein in Metal-Free, Intermediate and Metal-Bound States from Candidatus Liberibacter Asiaticus.
J.Struct.Biol., 189, 2015
|
|
4UDO
 
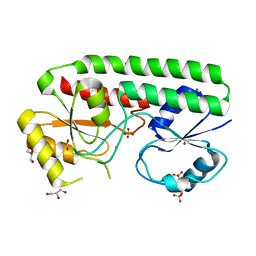 | | structure of Mn-bound periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Sharma, N, Selvakumar, P, Kumar, P, Sharma, A.K. | | Deposit date: | 2014-12-10 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of a Periplasmic Solute Binding Protein in Metal-Free, Intermediate and Metal-Bound States from Candidatus Liberibacter Asiaticus.
J.Struct.Biol., 189, 2015
|
|
4CL2
 
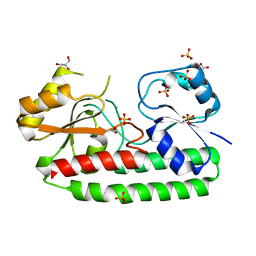 | | structure of periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Sharma, N, Selvakumar, P, Bhose, S, Ghosh, D.K, Kumar, P, Sharma, A.K. | | Deposit date: | 2014-01-11 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of a Periplasmic Solute Binding Protein in Metal-Free, Intermediate and Metal-Bound States from Candidatus Liberibacter Asiaticus.
J.Struct.Biol., 189, 2015
|
|
