3CG6
 
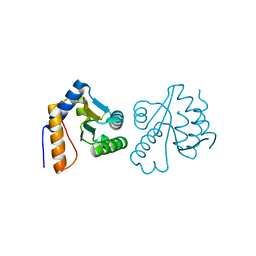 | | Crystal structure of Gadd45 gamma | | Descriptor: | Growth arrest and DNA-damage-inducible 45 gamma | | Authors: | Schrag, J.D, Jiralerspong, S, Banville, M, Jaramillo, M.L, O'Connor-McCourt, M.D. | | Deposit date: | 2008-03-05 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure and dimerization interface of GADD45gamma.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1THG
 
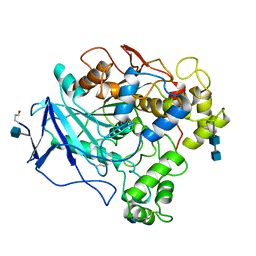 | | 1.8 ANGSTROMS REFINED STRUCTURE OF THE LIPASE FROM GEOTRICHUM CANDIDUM | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schrag, J.D, Cygler, M. | | Deposit date: | 1992-07-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 A refined structure of the lipase from Geotrichum candidum.
J.Mol.Biol., 230, 1993
|
|
2LIP
 
 | |
1JHN
 
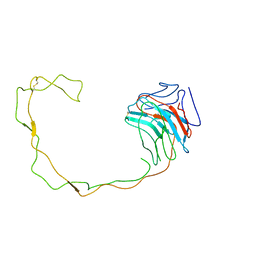 | | Crystal Structure of the Lumenal Domain of Calnexin | | Descriptor: | CALCIUM ION, calnexin | | Authors: | Schrag, J.D, Bergeron, J.M, Li, Y, Borisova, S, Hahn, M, Thomas, D.Y, Cygler, M. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of calnexin, an ER chaperone involved in quality control of protein folding.
Mol.Cell, 8, 2001
|
|
3CPT
 
 | | MP1-p14 Scaffolding complex | | Descriptor: | Mitogen-activated protein kinase kinase 1-interacting protein 1, Mitogen-activated protein-binding protein-interacting protein | | Authors: | Schrag, J.D, Cygler, M, Munger, C, Magloire, A. | | Deposit date: | 2008-04-01 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular dynamics-solvated interaction energy studies of protein-protein interactions: the MP1-p14 scaffolding complex.
J.Mol.Biol., 379, 2008
|
|
3IA2
 
 | | Pseudomonas fluorescens esterase complexed to the R-enantiomer of a sulfonate transition state analog | | Descriptor: | (2R)-butane-2-sulfonate, Arylesterase, GLYCEROL, ... | | Authors: | Schrag, J.D, Kazlauskas, R.J, Jiang, Y, Morley, K. | | Deposit date: | 2009-07-13 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Different active-site loop orientation in serine hydrolases versus acyltransferases.
Chembiochem, 12, 2011
|
|
2ZL1
 
 | | MP1-p14 Scaffolding complex | | Descriptor: | Mitogen-activated protein kinase kinase 1-interacting protein 1, Mitogen-activated protein-binding protein-interacting protein | | Authors: | Schrag, J.D, Cygler, M, Munger, C, Magloire, A. | | Deposit date: | 2008-04-02 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular dynamics-solvated interaction energy studies of protein-protein interactions: the MP1-p14 scaffolding complex.
J.Mol.Biol., 379, 2008
|
|
3HEA
 
 | | The L29P/L124I mutation of Pseudomonas fluorescens esterase | | Descriptor: | Arylesterase, ETHYL ACETATE, GLYCEROL, ... | | Authors: | Kazlauskas, R.J, Schrag, J.D, Cheeseman, J.D, Morley, K.L. | | Deposit date: | 2009-05-08 | | Release date: | 2010-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Switching catalysis from hydrolysis to perhydrolysis in Pseudomonas fluorescens esterase.
Biochemistry, 49, 2010
|
|
1KSK
 
 | | STRUCTURE OF RSUA | | Descriptor: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URACIL | | Authors: | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2002-01-13 | | Release date: | 2002-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
1VA4
 
 | | Pseudomonas fluorescens aryl esterase | | Descriptor: | Arylesterase, GLYCEROL | | Authors: | Cheeseman, J.D, Tocilj, A, Park, S, Schrag, J.D, Kazlauskas, R.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structure of an aryl esterase from Pseudomonas fluorescens.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3PT5
 
 | | Crystal structure of NanS | | Descriptor: | NANS (YJHS), A 9-O-acetyl N-acetylneuraminic acid esterase | | Authors: | Ruane, K.M, Rangarajan, E.S, Proteau, A, Schrag, J.D, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-12-02 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and enzymatic characterization of NanS (YjhS), a 9-O-Acetyl N-acetylneuraminic acid esterase from Escherichia coli O157:H7.
Protein Sci., 20, 2011
|
|
1YNI
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | N~2~-(3-CARBOXYPROPANOYL)-L-ARGININE, POTASSIUM ION, Succinylarginine Dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
2H8L
 
 | |
1EEA
 
 | | Acetylcholinesterase | | Descriptor: | PROTEIN (ACETYLCHOLINESTERASE) | | Authors: | Raves, M.L, Giles, K, Schrag, J.D, Schmid, M.F, Phillips Jr, G.N, Wah, C, Howard, A.J, Silman, I, Sussman, J.L. | | Deposit date: | 1999-01-26 | | Release date: | 1999-02-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Quaternary Structure of Tetrameric Acetylcholinesterase
Structure and Function of Cholinesterases and Related Proteins, 1998
|
|
1YS2
 
 | | Burkholderia cepacia lipase complexed with hexylphosphonic acid (S) 2-methyl-3-phenylpropyl ester | | Descriptor: | CALCIUM ION, HEXYLPHOSPHONIC ACID (S)-2-METHYL-3-PHENYLPROPYL ESTER, Lipase | | Authors: | Mezzetti, A, Schrag, J.D, Cheong, C.S, Kazlauskas, R.J. | | Deposit date: | 2005-02-06 | | Release date: | 2005-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mirror-Image Packing in Enantiomer Discrimination Molecular Basis for the Enantioselectivity of B.cepacia Lipase toward 2-Methyl-3-Phenyl-1-Propanol.
Chem.Biol., 12, 2005
|
|
1YS1
 
 | | Burkholderia cepacia lipase complexed with hexylphosphonic acid (R)-2-methyl-3-phenylpropyl ester | | Descriptor: | CALCIUM ION, HEXYLPHOSPHONIC ACID (R)-2-METHYL-3-PHENYLPROPYL ESTER, Lipase | | Authors: | Mezzetti, A, Schrag, J.D, Cheong, C.S, Kazlauskas, R.J. | | Deposit date: | 2005-02-06 | | Release date: | 2005-05-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mirror-Image Packing in Enantiomer Discrimination Molecular Basis for the Enantioselectivity of B.cepacia Lipase toward 2-Methyl-3-Phenyl-1-Propanol.
Chem.Biol., 12, 2005
|
|
1YNF
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | POTASSIUM ION, Succinylarginine dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
1YNH
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | N~2~-(3-CARBOXYPROPANOYL)-L-ORNITHINE, POTASSIUM ION, Succinylarginine Dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M. | | Deposit date: | 2005-01-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
1ZPS
 
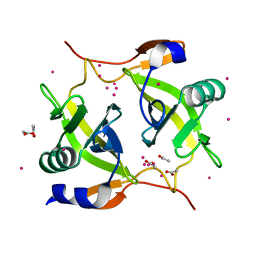 | | Crystal structure of Methanobacterium thermoautotrophicum phosphoribosyl-AMP cyclohydrolase HisI | | Descriptor: | ACETIC ACID, CADMIUM ION, Phosphoribosyl-AMP cyclohydrolase | | Authors: | Sivaraman, J, Myers, R.S, Boju, L, Sulea, T, Cygler, M, Davisson, V.J, Schrag, J.D, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Methanobacterium thermoautotrophicum Phosphoribosyl-AMP Cyclohydrolase HisI.
Biochemistry, 44, 2005
|
|
1Q18
 
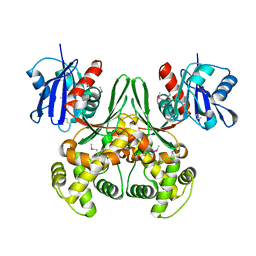 | | Crystal structure of E.coli glucokinase (Glk) | | Descriptor: | Glucokinase | | Authors: | Lunin, V.V, Li, Y, Schrag, J.D, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-07-18 | | Release date: | 2004-07-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose.
J.Bacteriol., 186, 2004
|
|
1SSL
 
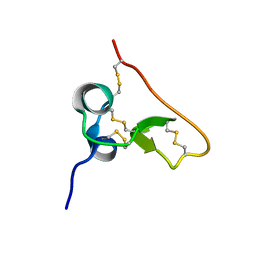 | | Solution structure of the PSI domain from the Met receptor | | Descriptor: | Hepatocyte growth factor receptor | | Authors: | Kozlov, G, Perreault, A, Schrag, J.D, Cygler, M, Gehring, K, Ekiel, I. | | Deposit date: | 2004-03-24 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Insights into function of PSI domains from structure of the Met receptor PSI domain.
Biochem.Biophys.Res.Commun., 321, 2004
|
|
1SZ2
 
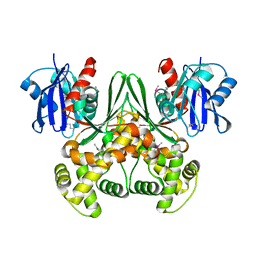 | | Crystal structure of E. coli glucokinase in complex with glucose | | Descriptor: | Glucokinase, beta-D-glucopyranose | | Authors: | Lunin, V.V, Li, Y, Schrag, J.D, Iannuzzi, P, Matte, A, Cygler, M. | | Deposit date: | 2004-04-02 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose
J.Bacteriol., 186, 2004
|
|
1FC4
 
 | | 2-AMINO-3-KETOBUTYRATE COA LIGASE | | Descriptor: | 2-AMINO-3-KETOBUTYRATE CONENZYME A LIGASE, 2-AMINO-3-KETOBUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schmidt, A, Matte, A, Li, Y, Sivaraman, J, Larocque, R, Schrag, J.D, Smith, C, Sauve, V, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2000-07-17 | | Release date: | 2001-05-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of 2-amino-3-ketobutyrate CoA ligase from Escherichia coli complexed with a PLP-substrate intermediate: inferred reaction mechanism.
Biochemistry, 40, 2001
|
|
1IJI
 
 | | Crystal Structure of L-Histidinol Phosphate Aminotransferase with PLP | | Descriptor: | Histidinol Phosphate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sivaraman, J, Li, Y, Larocque, R, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2001-04-26 | | Release date: | 2001-08-29 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of histidinol phosphate aminotransferase (HisC) from Escherichia coli, and its covalent complex with pyridoxal-5'-phosphate and l-histidinol phosphate.
J.Mol.Biol., 311, 2001
|
|
1KAR
 
 | | L-HISTIDINOL DEHYDROGENASE (HISD) STRUCTURE COMPLEXED WITH HISTAMINE (INHIBITOR), ZINC AND NAD (COFACTOR) | | Descriptor: | HISTAMINE, Histidinol dehydrogenase, ZINC ION | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J.D, Cygler, M. | | Deposit date: | 2001-11-02 | | Release date: | 2002-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
