6P0Y
 
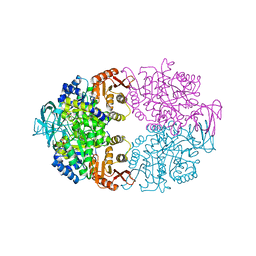 | | Cryptosporidium parvum pyruvate kinase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2019-05-17 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An overview of structure, function, and regulation of pyruvate kinases.
Protein Sci., 28, 2019
|
|
4IRB
 
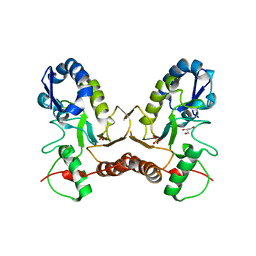 | | Crystal Structure of Vaccinia Virus Uracil DNA Glycosylase Mutant del171-172D4 | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Schormann, N, Zhukovskaya, N, Sartmatova, D, Nuth, M, Ricciardi, R.P, Chattopadhyay, D. | | Deposit date: | 2013-01-14 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutations at the dimer interface affect both function and structure of the Vaccinia virus uracil DNA glycosylase
To be Published
|
|
1MXF
 
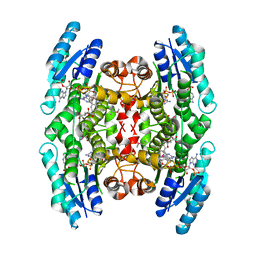 | | Crystal Structure of Inhibitor Complex of Putative Pteridine Reductase 2 (PTR2) from Trypanosoma cruzi | | Descriptor: | METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 2 | | Authors: | Schormann, N, Pal, B, Senkovich, O, Carson, M, Howard, A, Smith, C, Delucas, L, Chattopadhyay, D. | | Deposit date: | 2002-10-02 | | Release date: | 2003-10-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Trypanosoma cruzi pteridine reductase 2 in complex with a substrate and an inhibitor.
J.Struct.Biol., 152, 2005
|
|
1B0W
 
 | |
1MXH
 
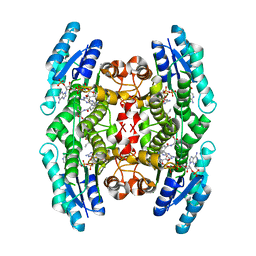 | | Crystal Structure of Substrate Complex of Putative Pteridine Reductase 2 (PTR2) from Trypanosoma cruzi | | Descriptor: | DIHYDROFOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 2 | | Authors: | Schormann, N, Pal, B, Senkovich, O, Carson, M, Howard, A, Smith, C, Delucas, L, Chattopadhyay, D. | | Deposit date: | 2002-10-02 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Trypanosoma cruzi pteridine reductase 2 in complex with a substrate and an inhibitor.
J.Struct.Biol., 152, 2005
|
|
3KJS
 
 | | Crystal Structure of T. cruzi DHFR-TS with 3 high affinity DHFR inhibitors: DQ1 inhibitor complex | | Descriptor: | 1,2-ETHANEDIOL, Dihydrofolate reductase-thymidylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2009-11-03 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthesis and characterization of potent inhibitors of Trypanosoma cruzi dihydrofolate reductase.
Bioorg.Med.Chem., 18, 2010
|
|
2B34
 
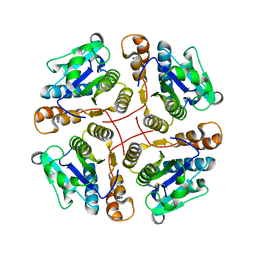 | | Structure of MAR1 Ribonuclease from Caenorhabditis elegans | | Descriptor: | MAR1 Ribonuclease | | Authors: | Schormann, N, Karpova, E, Li, S, Symersky, J, Zhang, Y, Lu, S, Zhou, Q, Lin, G, Cao, Z, Luo, M, Qiu, S, Luan, C.-H, Luo, D, Huang, W, Shang, Q, McKinstry, A, An, J, Tsao, J, Carson, M, Stinnett, M, Chen, Y, Johnson, D, Gary, R, Arabshahi, A, Bunzel, R, Bray, T, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-09-19 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structure of MAR1 Ribonuclease from Caenorhabditis elegans
To be Published
|
|
7L0O
 
 | |
7LGR
 
 | |
2TRH
 
 | |
2TRY
 
 | |
6UBV
 
 | |
6TZL
 
 | |
2GP4
 
 | |
6Q2K
 
 | |
6Q2L
 
 | |
3NHE
 
 | |
4LZB
 
 | | Uracil binding pocket in Vaccinia virus uracil DNA glycosylase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the uracil complex of Vaccinia virus uracil DNA glycosylase.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1BZD
 
 | |
1BZ8
 
 | |
1BZE
 
 | |
1BRE
 
 | | IMMUNOGLOBULIN LIGHT CHAIN PROTEIN | | Descriptor: | BENCE-JONES KAPPA I PROTEIN BRE | | Authors: | Schormann, N, Benson, M.D. | | Deposit date: | 1995-07-19 | | Release date: | 1995-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tertiary structure of an amyloid immunoglobulin light chain protein: a proposed model for amyloid fibril formation.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
4QCB
 
 | | Protein-DNA complex of Vaccinia virus D4 with double-stranded non-specific DNA | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*C)-3', GLYCEROL, Uracil-DNA glycosylase | | Authors: | Schormann, N, Banerjee, S, Ricciardi, R, Chattopadhyay, D. | | Deposit date: | 2014-05-09 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Binding of undamaged double stranded DNA to vaccinia virus uracil-DNA Glycosylase.
BMC Struct. Biol., 15, 2015
|
|
3CL9
 
 | | Structure of bifunctional TcDHFR-TS in complex with MTX | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase (DHFR-TS), ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2008-03-18 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function.
Proteins, 73, 2008
|
|
3CLB
 
 | | Structure of bifunctional TcDHFR-TS in complex with TMQ | | Descriptor: | 1,2-ETHANEDIOL, DHFR-TS, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2008-03-18 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function.
Proteins, 73, 2008
|
|
