3RKP
 
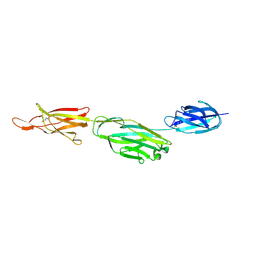 | | Crystal structure of BcpA*(D312A), the major pilin subunit of Bacillus cereus | | Descriptor: | Collagen adhesion protein | | Authors: | Hendrickx, A.P, Poor, C.B, Jureller, J.E, Budzik, J.M, He, C, Schneewind, O. | | Deposit date: | 2011-04-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | Isopeptide bonds of the major pilin protein BcpA influence pilus structure and bundle formation on the surface of Bacillus cereus.
Mol.Microbiol., 85, 2012
|
|
3KPT
 
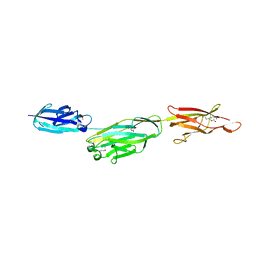 | | Crystal structure of BcpA, the major pilin subunit of Bacillus cereus | | Descriptor: | CALCIUM ION, Collagen adhesion protein | | Authors: | Poor, C.B, Budzik, J.M, Schneewind, O, He, C. | | Deposit date: | 2009-11-16 | | Release date: | 2009-11-24 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Intramolecular amide bonds stabilize pili on the surface of bacilli.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1QWZ
 
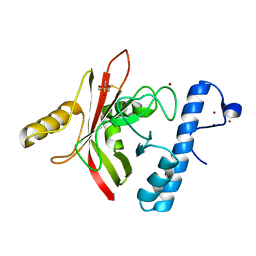 | | Crystal structure of Sortase B from S. aureus complexed with MTSET | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, NICKEL (II) ION, NPQTN specific sortase B, ... | | Authors: | Zong, Y, Mazmanian, S.K, Schneewind, O, Narayana, S.V. | | Deposit date: | 2003-09-03 | | Release date: | 2004-04-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of sortase B, a cysteine transpeptidase that tethers surface protein to the Staphylococcus aureus cell wall
Structure, 12, 2004
|
|
1QX6
 
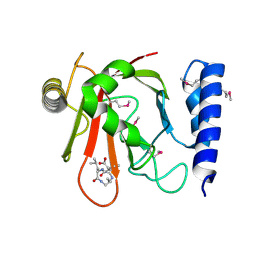 | | Crystal structure of Sortase B complexed with E-64 | | Descriptor: | N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, NPQTN specific sortase B | | Authors: | Zong, Y, Mazmanian, S.K, Schneewind, O, Narayana, S.V. | | Deposit date: | 2003-09-04 | | Release date: | 2004-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of sortase B, a cysteine transpeptidase that tethers surface protein to the Staphylococcus aureus cell wall
Structure, 12, 2004
|
|
1QXA
 
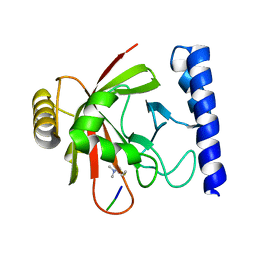 | | Crystal structure of Sortase B complexed with Gly3 | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, NPQTN specific sortase B, peptide GLY-GLY-GLY | | Authors: | Zong, Y, Mazmanian, S.K, Schneewind, O, Narayana, S.V. | | Deposit date: | 2003-09-05 | | Release date: | 2004-04-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of sortase B, a cysteine transpeptidase that tethers surface protein to the Staphylococcus aureus cell wall
Structure, 12, 2004
|
|
1XBW
 
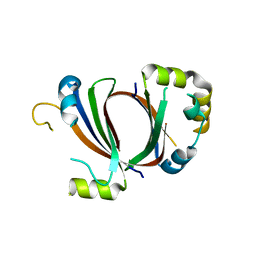 | | 1.9A Crystal Structure of the protein isdG from Staphylococcus aureus aureus, Structural genomics, MCSG | | Descriptor: | hypothetical protein isdG | | Authors: | Zhang, R, Wu, R, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-31 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases.
J.Biol.Chem., 280, 2005
|
|
1IJA
 
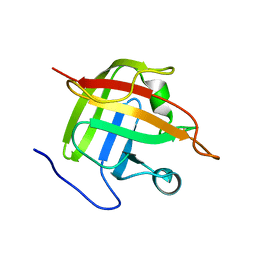 | | Structure of Sortase | | Descriptor: | Sortase | | Authors: | Ilangovan, U, Ton-That, H, Iwahara, J, Schneewind, O, Clubb, R.T. | | Deposit date: | 2001-04-25 | | Release date: | 2001-05-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5V4F
 
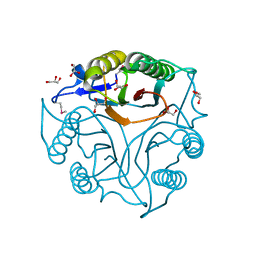 | | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfB from Yersinia pestis | | Descriptor: | GLYCEROL, Putative translational inhibitor protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Krishnan, A, Babnigg, G, Schneewind, O, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-03-09 | | Release date: | 2017-04-05 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfB from Yersinia pestis
To Be Published
|
|
5V4D
 
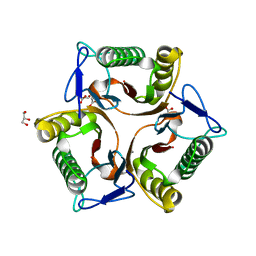 | | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfA from Yersinia pestis | | Descriptor: | ACETIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Krishnan, A, Babnigg, G, Schneewind, O, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-03-09 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfA from Yersinia pestis
To Be Published
|
|
2BV6
 
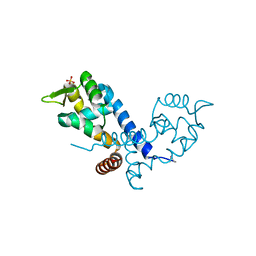 | | Crystal structure of MgrA, a global regulator and major virulence determinant in Staphylococcus aureus | | Descriptor: | HTH-TYPE TRANSCRIPTIONAL REGULATOR MGRA, SULFATE ION | | Authors: | Chen, P.R, Bae, T, Williams, W.A, Duguid, E.M, Rice, P.A, Schneewind, O, He, C. | | Deposit date: | 2005-06-22 | | Release date: | 2006-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An Oxidation-Sensing Mechanism is Used by the Global Regulator Mgra in Staphylococcus Aureus.
Nat.Chem.Biol., 2, 2006
|
|
2AP3
 
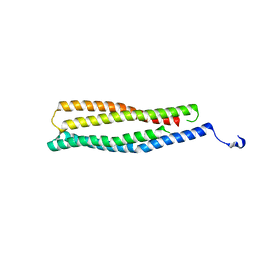 | | 1.6 A Crystal Structure of a Conserved Protein of Unknown Function from Staphylococcus aureus | | Descriptor: | conserved hypothetical protein | | Authors: | Zhang, R, Zhou, M, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-15 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6A crystal structure of a conserved hypothetical protein from Staphylococcus aureus MW2
To be Published
|
|
2OQW
 
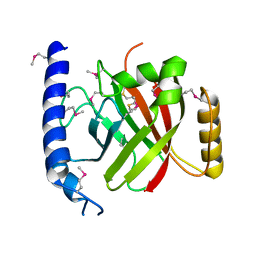 | | The crystal structure of sortase B from B.anthracis in complex with AAEK1 | | Descriptor: | Sortase B | | Authors: | Wu, R, Zhang, R, Marresso, A.W, Schneewind, O, Joachimiak, A. | | Deposit date: | 2007-02-01 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activation of inhibitors by sortase triggers irreversible modification of the active site.
J.Biol.Chem., 282, 2007
|
|
2OQZ
 
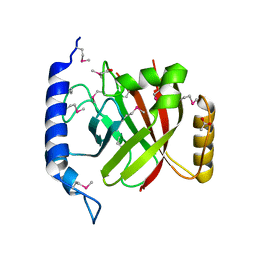 | | The crystal structure of sortase B from B.anthracis in complex with AAEK2 | | Descriptor: | ACETIC ACID, Sortase B | | Authors: | Wu, R, Zhang, R, Maresso, A.W, Schneewind, O, Joachimiak, A. | | Deposit date: | 2007-02-01 | | Release date: | 2007-06-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Activation of inhibitors by sortase triggers irreversible modification of the active site.
J.Biol.Chem., 282, 2007
|
|
3PYW
 
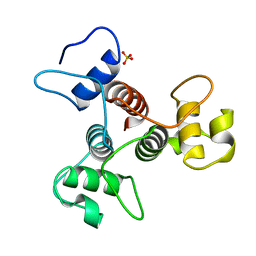 | | The structure of the SLH domain from B. anthracis surface array protein at 1.8A | | Descriptor: | S-layer protein sap, SULFATE ION | | Authors: | Zhang, R, Wilton, R, Kern, J, Joachimiak, A, Schneewind, O, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-12-13 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Surface Layer Homology (SLH) Domains from Bacillus anthracis Surface Array Protein.
J.Biol.Chem., 286, 2011
|
|
3NGG
 
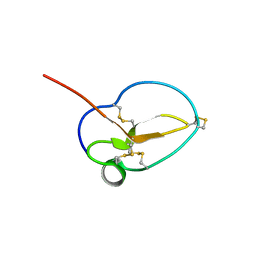 | | X-ray Structure of Omwaprin | | Descriptor: | Omwaprin-a | | Authors: | Banigan, J.R, Mandal, K, Sawaya, M.R, Thammavongsa, V, Hendrickx, A.P.A, Schneewind, O, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2010-06-11 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Determination of the X-ray structure of the snake venom protein omwaprin by total chemical synthesis and racemic protein crystallography.
Protein Sci., 19, 2010
|
|
1SQE
 
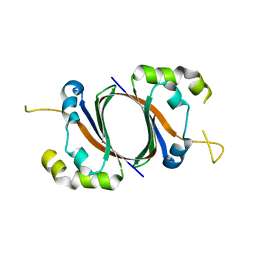 | | 1.5A Crystal Structure Of the protein PG130 from Staphylococcus aureus, Structural genomics | | Descriptor: | hypothetical protein PG130 | | Authors: | Zhang, R, Wu, R, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases
J.Biol.Chem., 280, 2005
|
|
1T2W
 
 | | Crystal Structure of Sortase A in Complex with a LPETG peptide | | Descriptor: | Class A sortase SrtA, Peptide LEU-PRO-GLU-THR-GLY | | Authors: | Zong, Y, Bice, T.W, Ton-That, H, Schneewind, O, Narayana, S.V. | | Deposit date: | 2004-04-23 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Staphylococcus aureus sortase A and its substrate complex
J.Biol.Chem., 279, 2004
|
|
2W5R
 
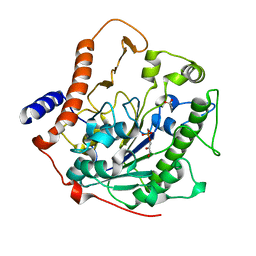 | | Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. | | Descriptor: | (2R)-2,3-dihydroxypropyl phosphate, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Lu, D, Wormann, M.E, Zhang, X, Schneewind, O, Grundling, A, Freemont, P.S. | | Deposit date: | 2008-12-11 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Mechanism of Lipoteichoic Acid Synthesis by Staphylococcus Aureus Ltas.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2W5S
 
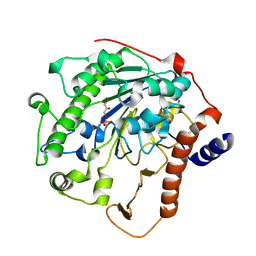 | | Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. | | Descriptor: | (2R)-2,3-dihydroxypropyl phosphate, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Lu, D, Wormann, M.E, Zhang, X, Schneewind, O, Grundling, A, Freemont, P.S. | | Deposit date: | 2008-12-11 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Mechanism of Lipoteichoic Acid Synthesis by Staphylococcus Aureus Ltas.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2W5T
 
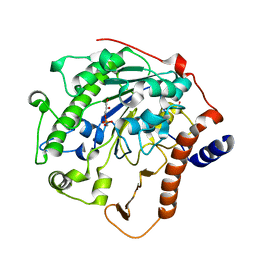 | | Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. | | Descriptor: | (2R)-2,3-dihydroxypropyl phosphate, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Lu, D, Wormann, M.E, Zhang, X, Schneewind, O, Grundling, A, Freemont, P.S. | | Deposit date: | 2008-12-11 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Mechanism of Lipoteichoic Acid Synthesis by Staphylococcus Aureus Ltas.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1T2P
 
 | | Crystal structure of Sortase A from Staphylococcus aureus | | Descriptor: | sortase | | Authors: | Zong, Y, Bice, T.W, Ton-That, H, Schneewind, O, Narayana, S.V. | | Deposit date: | 2004-04-22 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURES OF STAPHYLOCOCCUS AUREUS SORTASE A AND ITS SUBSTRATE COMPLEX
J.Biol.Chem., 279, 2004
|
|
1T2O
 
 | | Crystal structure of Se-SrtA, C184-Ala | | Descriptor: | sortase | | Authors: | Zong, Y, Bice, T.W, Ton-That, H, Schneewind, O, Narayana, S.V. | | Deposit date: | 2004-04-22 | | Release date: | 2004-09-07 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of Staphylococcus aureus sortase A and its substrate complex
J.Biol.Chem., 279, 2004
|
|
7TE5
 
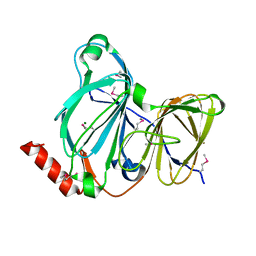 | | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK from Yersinia pestis | | Descriptor: | MAGNESIUM ION, Pirin family protein Yhak | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-04 | | Release date: | 2022-01-12 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK from Yersinia pestis
To Be Published
|
|
7TEM
 
 | | Crystal Structure of the Putative Exported Protein YPO2471 from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-05 | | Release date: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Putative Exported Protein YPO2471 from Yersinia pestis
To Be Published
|
|
7TG5
 
 | | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK in the Presence of Fe Ion from Yersinia pestis | | Descriptor: | CHLORIDE ION, FE (III) ION, Pirin family protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-07 | | Release date: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK in the presence of Fe ion from Yersinia pestis
To Be Published
|
|
