1MUX
 
 | | SOLUTION NMR STRUCTURE OF CALMODULIN/W-7 COMPLEX: THE BASIS OF DIVERSITY IN MOLECULAR RECOGNITION, 30 STRUCTURES | | Descriptor: | CALCIUM ION, CALMODULIN, N-(6-AMINOHEXYL)-5-CHLORO-1-NAPHTHALENESULFONAMIDE | | Authors: | Osawa, M, Swindells, M.B, Tanikawa, J, Tanaka, T, Mase, T, Furuya, T, Ikura, M. | | Deposit date: | 1997-09-06 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calmodulin-W-7 complex: the basis of diversity in molecular recognition.
J.Mol.Biol., 276, 1998
|
|
2RQH
 
 | | Structure of GSPT1/ERF3A-PABC | | Descriptor: | G1 to S phase transition 1, Polyadenylate-binding protein 1 | | Authors: | Osawa, M, Nakanishi, T, Hosoda, N, Uchida, S, Hoshino, T, Katada, I, Shimada, I. | | Deposit date: | 2009-05-08 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic Translation Termination Factor Gspt/Erf3 Recognizes Pabp with Chemical Exchange Using Two Overlapping Motifs
To be Published
|
|
2RQG
 
 | | Structure of GSPT1/ERF3A-PABC | | Descriptor: | G1 to S phase transition 1, Polyadenylate-binding protein 1 | | Authors: | Osawa, M, Nakanishi, T, Hosoda, N, Uchida, S, Hoshino, T, Katada, I, Shimada, I. | | Deposit date: | 2009-05-08 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic Translation Termination Factor Gspt/Erf3 Recognizes Pabp with Chemical Exchange Using Two Overlapping Motifs
To be Published
|
|
1CKK
 
 | | CALMODULIN/RAT CA2+/CALMODULIN DEPENDENT PROTEIN KINASE FRAGMENT | | Descriptor: | CALCIUM ION, Calcium/calmodulin-dependent protein kinase kinase 1, Calmodulin-1 | | Authors: | Osawa, M, Tokumitsu, H, Swindells, M.B, Kurihara, H, Orita, M, Shibanuma, T, Furuya, T, Ikura, M. | | Deposit date: | 1998-11-20 | | Release date: | 1999-09-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase.
Nat.Struct.Biol., 6, 1999
|
|
7W97
 
 | | Crystal Structure of the CYP102A1 (P450BM3) Heme Domain with N-Hexadecanoyl-L-Homoserine | | Descriptor: | (2~{S})-2-(hexadecanoylamino)-4-oxidanyl-butanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Karasawa, M, Stanfield, J.K, Kasai, C, Sugimoto, H, Shoji, O. | | Deposit date: | 2021-12-09 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the CYP102A1 (P450BM3) Heme Domain with N-Hexadecanoyl-L-Homoserine
To Be Published
|
|
7W9D
 
 | | Crystal Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 (P450BM3) Heme Domain with N-Hexadecanoyl-L-Homoserine | | Descriptor: | (2~{S})-2-(hexadecanoylamino)-4-oxidanyl-butanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Karasawa, M, Stanfield, J.K, Kasai, C, Sugimoto, H, Shoji, O. | | Deposit date: | 2021-12-09 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 (P450BM3) Heme Domain with N-Hexadecanoyl-L-Homoserine
To Be Published
|
|
7W9J
 
 | | Crystal Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 (P450BM3) Heme Domain with N-Dodecanoyl-L-Homoserine Lactone | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, N-[(3S)-2-oxooxolan-3-yl]dodecanamide, ... | | Authors: | Karasawa, M, Stanfield, J.K, Kasai, C, Sugimoto, H, Shoji, O. | | Deposit date: | 2021-12-09 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 (P450BM3) Heme Domain with N-Dodecanoyl-L-Homoserine Lactone
To Be Published
|
|
7DAN
 
 | |
5ZZ4
 
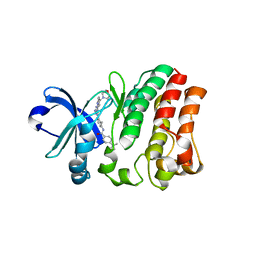 | | Crystal structure of bruton's tyrosine kinase in complex with inhibitor 2e | | Descriptor: | N-[3-(4-amino-6-{[4-(morpholine-4-carbonyl)phenyl]amino}-1,3,5-triazin-2-yl)-2-methylphenyl]-4-tert-butylbenzamide, Tyrosine-protein kinase BTK | | Authors: | Kawahata, W, Asami, T, Irie, T, Kiyoi, T, Taniguchi, H, Asamitsu, Y, Inoue, T, Miyake, T, Sawa, M. | | Deposit date: | 2018-05-30 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design and Synthesis of Novel Amino-triazine Analogues as Selective Bruton's Tyrosine Kinase Inhibitors for Treatment of Rheumatoid Arthritis.
J. Med. Chem., 61, 2018
|
|
5Y90
 
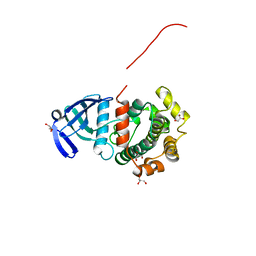 | | MAP2K7 mutant -C218S | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7, GLYCEROL | | Authors: | Kinoshita, T, Hashimoto, T, Sogabe, Y, Matsumoto, T, Sawa, M, Fukada, H. | | Deposit date: | 2017-08-22 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution structure discloses the potential for allosteric regulation of mitogen-activated protein kinase kinase 7
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5B2M
 
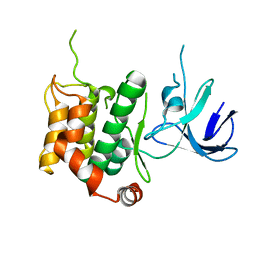 | | A crucial role of Cys218 in the stabilization of an unprecedented auto-inhibition form of MAP2K7 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Sogabe, Y, Hashimoto, T, Matsumoto, T, Kirii, Y, Sawa, M, Kinoshita, T. | | Deposit date: | 2016-01-19 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A crucial role of Cys218 in configuring an unprecedented auto-inhibition form of MAP2K7
Biochem.Biophys.Res.Commun., 473, 2016
|
|
5B2K
 
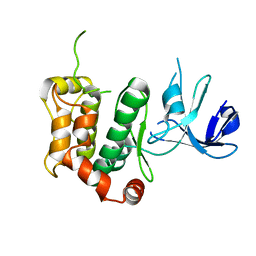 | | A crucial role of Cys218 in the stabilization of an unprecedented auto-inhibition form of MAP2K7 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Sogabe, Y, Hashimoto, T, Matsumoto, T, Kirii, Y, Sawa, M, Kinoshita, T. | | Deposit date: | 2016-01-19 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A crucial role of Cys218 in configuring an unprecedented auto-inhibition form of MAP2K7
Biochem.Biophys.Res.Commun., 473, 2016
|
|
5B2L
 
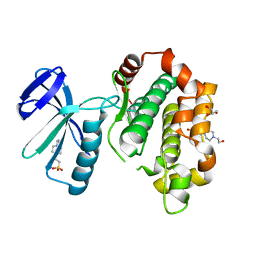 | | A crucial role of Cys218 in the stabilization of an unprecedented auto-inhibition form of MAP2K7 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Dual specificity mitogen-activated protein kinase kinase 7, GLYCEROL | | Authors: | Sogabe, Y, Hashimoto, T, Matsumoto, T, Kirii, Y, Sawa, M, Kinoshita, T. | | Deposit date: | 2016-01-19 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A crucial role of Cys218 in configuring an unprecedented auto-inhibition form of MAP2K7
Biochem.Biophys.Res.Commun., 473, 2016
|
|
8XBV
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the sticky end of the nucleosome | | Descriptor: | DNA (5'-D(P*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*A)-3'), DNA (5'-D(P*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*G)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.61 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBY
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the blunt end of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBU
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBX
 
 | | The cryo-EM structure of the RAD51 L2 loop bound to the linker DNA with the blunt end of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBT
 
 | | The cryo-EM structure of the octameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8YBK
 
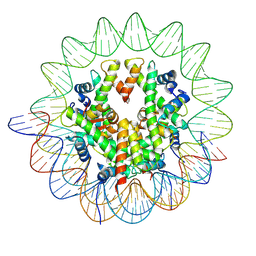 | | Cryo-EM structure of the human nucleosome containing the H3.1 E97K mutant | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kimura, T, Hirai, S, Kujirai, T, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Cryo-EM structure and biochemical analyses of the nucleosome containing the cancer-associated histone H3 mutation E97K.
Genes Cells, 29, 2024
|
|
8YBJ
 
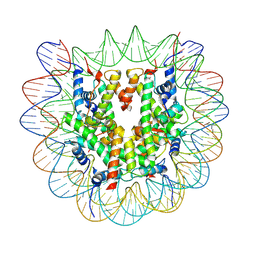 | | Cryo-EM structure of human nucleosome core particle composed of the Widom 601 DNA sequence | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kimura, T, Hirai, S, Kujirai, T, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structure and biochemical analyses of the nucleosome containing the cancer-associated histone H3 mutation E97K.
Genes Cells, 29, 2024
|
|
1IQ5
 
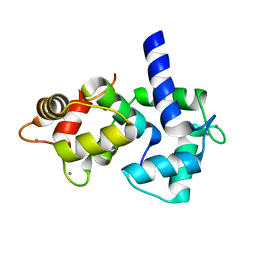 | | Calmodulin/nematode CA2+/Calmodulin dependent kinase kinase fragment | | Descriptor: | CA2+/CALMODULIN DEPENDENT KINASE KINASE, CALCIUM ION, CALMODULIN | | Authors: | Kurokawa, H, Osawa, M, Kurihara, H, Katayama, N, Tokumitsu, H, Swindells, M.B, Kainosho, M, Ikura, M. | | Deposit date: | 2001-06-14 | | Release date: | 2001-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Target-induced conformational adaptation of calmodulin revealed by the crystal structure of a complex with nematode Ca(2+)/calmodulin-dependent kinase kinase peptide
J.Mol.Biol., 312, 2001
|
|
4J70
 
 | | Yeast 20S proteasome in complex with the belactosin derivative 3e | | Descriptor: | Proteasome component C1, Proteasome component C11, Proteasome component C5, ... | | Authors: | Kawamura, S, Unno, Y, List, A, Tanaka, M, Sasaki, T, Arisawa, M, Asai, A, Groll, M, Shuto, S. | | Deposit date: | 2013-02-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Potent Proteasome Inhibitors Derived from the Unnatural cis-Cyclopropane Isomer of Belactosin A: Synthesis, Biological Activity, and Mode of Action.
J.Med.Chem., 56, 2013
|
|
8JL9
 
 | | Cryo-EM structure of the human nucleosome with scFv | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
8JLB
 
 | | Cryo-EM structure of the 145 bp human nucleosome containing H3.2 C110A mutant | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
