5JQZ
 
 | | Designed two-ring homotetramer at 3.8A resolution | | Descriptor: | De novo designed homotetramer | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Oberdorfer, G, Boyken, S.E, Chen, Z. | | Deposit date: | 2016-05-05 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.83 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J0I
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | Designed protein 2L6HC3_12 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J0K
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | designed protein 2L4HC2_23 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J0H
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | Design construct 2L6HC3_13 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J10
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | peptide design 2L4HC2_24 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J0L
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | designed protein 3L6HC2_2 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-03-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J0J
 
 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | designed protein 2L6HC3_6 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.256 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J73
 
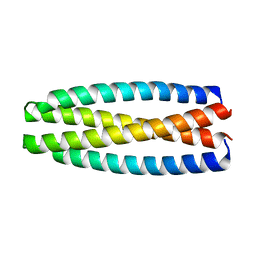 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | protein design 2L4HC2_9 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-04-05 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5J2L
 
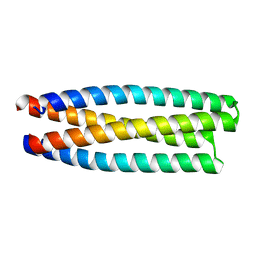 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | protein design 2L4HC2_11 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | Deposit date: | 2016-03-29 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
5IZS
 
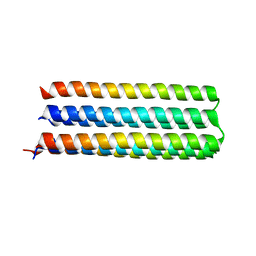 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | Descriptor: | Designed protein 5L6HC3_1 | | Authors: | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Oberdorfer, G. | | Deposit date: | 2016-03-25 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
6MFH
 
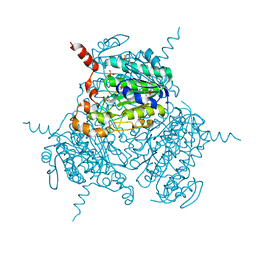 | | Mutated Uronate Dehydrogenase | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Mutated Uronate Dehydrogenase, PHOSPHATE ION | | Authors: | Sankaran, B, Pereira, J.H, Chan, V, Zwart, P.H, Wagschal, K. | | Deposit date: | 2018-09-11 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Chromohalobacter salixigens uronate dehydrogenase: Directed evolution for improved thermal stability and mutant CsUDH-inc X-ray crystal structure
Process Biochem, 2020
|
|
6V8E
 
 | |
5EIL
 
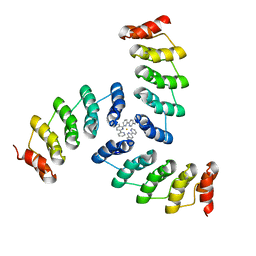 | | Computational design of a high-affinity metalloprotein homotrimer containing a metal chelating non-canonical amino acid | | Descriptor: | FE (III) ION, TRI-05 | | Authors: | Sankaran, B, Zwart, P.H, Mills, J.H, Pereira, J.H, Baker, D. | | Deposit date: | 2015-10-30 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Computational design of a homotrimeric metalloprotein with a trisbipyridyl core.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5K7V
 
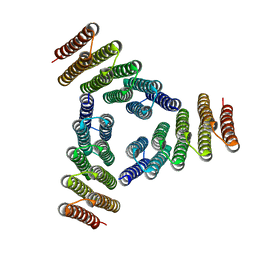 | | Computational Design of Self-Assembling Cyclic Protein Homooligomers | | Descriptor: | Designed protein HR00C3 | | Authors: | Sankaran, B, Zwart, P.H, Fallas, J.A, Pereira, J.H, Ueda, G, Baker, D. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.165 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
5KBA
 
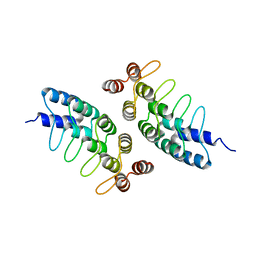 | | Computational Design of Self-Assembling Cyclic Protein Homooligomers | | Descriptor: | Designed protein ANK1C2 | | Authors: | Sankaran, B, Zwart, P.H, Fallas, J.A, Pereira, J.H, Ueda, G, Baker, D. | | Deposit date: | 2016-06-02 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
4W82
 
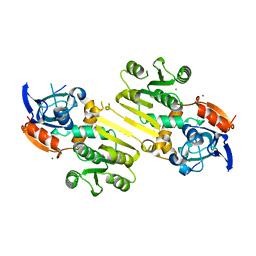 | | Enoyl-acyl carrier protein-reductase domain from human fatty acid synthase | | Descriptor: | CHLORIDE ION, Fatty acid synthase, MAGNESIUM ION, ... | | Authors: | Sippel, K.H, Vyas, N.K, Sankaran, B, Quiocho, F.A. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the human Fatty Acid synthase enoyl-acyl carrier protein-reductase domain complexed with triclosan reveals allosteric protein-protein interface inhibition.
J.Biol.Chem., 289, 2014
|
|
8T6C
 
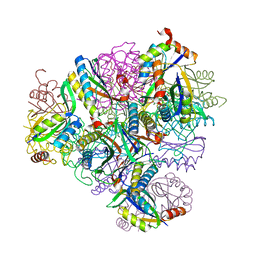 | | Crystal structure of T33-18.2: Deep-learning sequence design of co-assembling tetrahedron protein nanoparticles | | Descriptor: | T33-18.2 : A, T33-18.2 : B | | Authors: | Bera, A.K, de Haas, R.J, Kang, A, Sankaran, B, King, N.P. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Rapid and automated design of two-component protein nanomaterials using ProteinMPNN.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8T6E
 
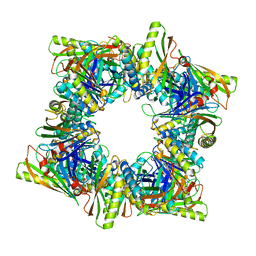 | | Crystal structure of T33-28.3: Deep-learning sequence design of co-assembling tetrahedron protein nanoparticles | | Descriptor: | T33-28.3: A, T33-28.3: B | | Authors: | Bera, A.K, de Haas, R.J, Kang, A, Sankaran, B, King, N.P. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Rapid and automated design of two-component protein nanomaterials using ProteinMPNN.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8T6N
 
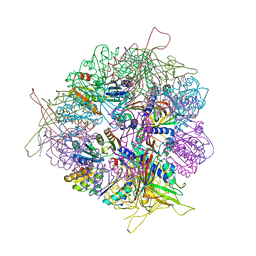 | | Crystal structure of T33-27.1: Deep-learning sequence design of co-assembling tetrahedron protein nanoparticles | | Descriptor: | T33-27.1 : A, T33-27.1 : B | | Authors: | Bera, A.K, de Haas, R.J, Kang, A, Sankaran, B, King, N.P. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Rapid and automated design of two-component protein nanomaterials using ProteinMPNN.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4WB4
 
 | | wt SA11 NSP4_CCD | | Descriptor: | CALCIUM ION, Non-structural glycoprotein NSP4 | | Authors: | Viskovska, M, Sastri, N.P, Hyser, J.M, Tanner, M.R, Horton, L.B, Sankaran, B, Prasad, B.V.V, Estes, M.K. | | Deposit date: | 2014-09-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Plasticity of the Coiled-Coil Domain of Rotavirus NSP4.
J.Virol., 88, 2014
|
|
4WBA
 
 | | Q/E mutant SA11 NSP4_CCD | | Descriptor: | GLYCEROL, Non-structural glycoprotein NSP4, PHOSPHATE ION | | Authors: | Viskovska, M, Sastri, N.P, Hyser, J.M, Tanner, M.R, Horton, L.B, Sankaran, B, Prasad, B.V.V, Estes, M.K. | | Deposit date: | 2014-09-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structural Plasticity of the Coiled-Coil Domain of Rotavirus NSP4.
J.Virol., 88, 2014
|
|
8SJ3
 
 | |
4S2L
 
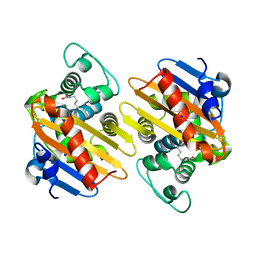 | | Crystal Structure of OXA-163 beta-lactamase | | Descriptor: | Beta-lactamase, SODIUM ION | | Authors: | Stojanoski, V, Liya, H, Palzkill, T.G, Prasad, B, Sankaran, B. | | Deposit date: | 2015-01-21 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for Different Substrate Profiles of Two Closely Related Class D beta-Lactamases and Their Inhibition by Halogens.
Biochemistry, 54, 2015
|
|
2PTM
 
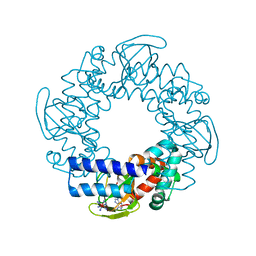 | | Structure and rearrangements in the carboxy-terminal region of SpIH channels | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, COBALT HEXAMMINE(III), Hyperpolarization-activated (Ih) channel | | Authors: | Flynn, G.E, Black, K.D, Islas, L.D, Sankaran, B, Zagotta, W.N. | | Deposit date: | 2007-05-08 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure and rearrangements in the carboxy-terminal region of SpIH channels.
Structure, 15, 2007
|
|
2Q0A
 
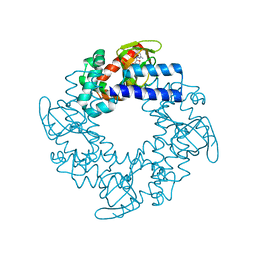 | | Structure and rearrangements in the carboxy-terminal region of SpIH channels | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Flynn, G.E, Black, K.D, Islas, L.D, Sankaran, B, Zagotta, W.N. | | Deposit date: | 2007-05-21 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and rearrangements in the carboxy-terminal region of SpIH channels.
Structure, 15, 2007
|
|
