2OM3
 
 | | High-resolution cryo-EM structure of Tobacco Mosaic Virus | | Descriptor: | Coat protein, Tobacco Mosaic Virus RNA | | Authors: | Sachse, C. | | Deposit date: | 2007-01-20 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | High-resolution electron microscopy of helical specimens: a fresh look at tobacco mosaic virus.
J.Mol.Biol., 371, 2007
|
|
5JM9
 
 | | Structure of S. cerevesiae mApe1 dodecamer | | Descriptor: | Vacuolar aminopeptidase 1 | | Authors: | Sachse, C, Bertipaglia, C. | | Deposit date: | 2016-04-28 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Higher-order assemblies of oligomeric cargo receptor complexes form the membrane scaffold of the Cvt vesicle.
Embo Rep., 17, 2016
|
|
7Q22
 
 | | cryo iDPC-STEM structure recorded with CSA 2.0 | | Descriptor: | Capsid protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Sachse, C, Leidl, M.L. | | Deposit date: | 2021-10-22 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Single-particle cryo-EM structures from iDPC-STEM at near-atomic resolution.
Nat.Methods, 19, 2022
|
|
7Q2Q
 
 | | cryo iDPC-STEM structure recorded with CSA 3.5 | | Descriptor: | Capsid protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Sachse, C, Leidl, M.L. | | Deposit date: | 2021-10-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Single-particle cryo-EM structures from iDPC-STEM at near-atomic resolution.
Nat.Methods, 19, 2022
|
|
7Q23
 
 | | cryo iDPC-STEM structure recorded with CSA 3.0 | | Descriptor: | Capsid protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Sachse, C, Leidl, M.L. | | Deposit date: | 2021-10-22 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Single-particle cryo-EM structures from iDPC-STEM at near-atomic resolution.
Nat.Methods, 19, 2022
|
|
7Q2S
 
 | | cryo iDPC-STEM structure recorded with CSA 4.5 | | Descriptor: | Capsid protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Sachse, C, Leidl, M.L. | | Deposit date: | 2021-10-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Single-particle cryo-EM structures from iDPC-STEM at near-atomic resolution.
Nat.Methods, 19, 2022
|
|
7Q2R
 
 | | cryo iDPC-STEM structure recorded with CSA 4.0 | | Descriptor: | Capsid protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Sachse, C, Leidl, M.L. | | Deposit date: | 2021-10-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Single-particle cryo-EM structures from iDPC-STEM at near-atomic resolution.
Nat.Methods, 19, 2022
|
|
4UF9
 
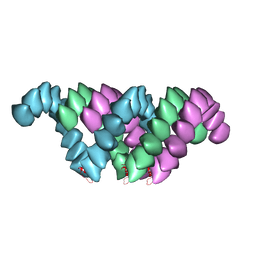 | | Electron cryo-microscopy structure of PB1-p62 type T filaments | | Descriptor: | SEQUESTOSOME-1 | | Authors: | Ciuffa, R, Lamark, T, Tarafder, A, Guesdon, A, Rybina, S, Hagen, W.J.H, Johansen, T, Sachse, C. | | Deposit date: | 2015-03-15 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.3 Å) | | Cite: | The Selective Autophagy Receptor P62 Forms a Flexible Filamentous Helical Scaffold.
Cell Rep., 11, 2015
|
|
9FN1
 
 | | 360 A Loki CHMP4-7 rods | | Descriptor: | Vps20/32/60-like protein (ESCRT-III) | | Authors: | Melnikov, N, Junglas, B, Halbi, G, Nachmias, D, Upcher, A, Zalk, R, Sachse, C, Bernheim-Groswasser, A, Elia, N. | | Deposit date: | 2024-06-07 | | Release date: | 2025-04-16 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | The Asgard archaeal ESCRT-III system remodels eukaryotic membranes, shedding light on the emergence of eukaryogenesis
To Be Published
|
|
7ZW6
 
 | | Oligomeric structure of SynDLP | | Descriptor: | Slr0869 protein | | Authors: | Gewehr, L, Junglas, B, Jilly, R, Franz, J, Wenyu, E.Z, Weidner, T, Bonn, M, Sachse, C, Schneider, D. | | Deposit date: | 2022-05-18 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | SynDLP is a dynamin-like protein of Synechocystis sp. PCC 6803 with eukaryotic features.
Nat Commun, 14, 2023
|
|
6ZH3
 
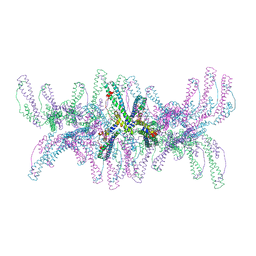 | |
5JM0
 
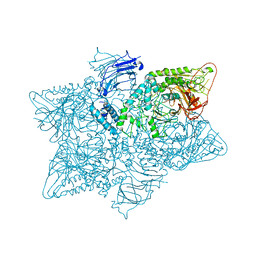 | | Structure of the S. cerevisiae alpha-mannosidase 1 | | Descriptor: | Alpha-mannosidase,Alpha-mannosidase,Alpha-mannosidase | | Authors: | Schneider, S, Kosinski, J, Jakobi, A.J, Hagen, W.J.H, Sachse, C. | | Deposit date: | 2016-04-28 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Higher-order assemblies of oligomeric cargo receptor complexes form the membrane scaffold of the Cvt vesicle.
Embo Rep., 17, 2016
|
|
7ABK
 
 | | Helical structure of PspA | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Huber, S.T, Mann, D, Heidler, T, Clarke, M, Schneider, D, Sachse, C. | | Deposit date: | 2020-09-07 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | PspA adopts an ESCRT-III-like fold and remodels bacterial membranes.
Cell, 184, 2021
|
|
2W6D
 
 | | BACTERIAL DYNAMIN-LIKE PROTEIN LIPID TUBE BOUND | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DYNAMIN FAMILY PROTEIN, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Low, H.H, Sachse, C, Amos, L.A, Lowe, J. | | Deposit date: | 2008-12-18 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of a Bacterial Dynamin-Like Protein Lipid Tube Provides a Mechanism for Assembly and Membrane Curving.
Cell(Cambridge,Mass.), 139, 2009
|
|
5JM6
 
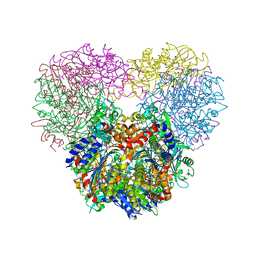 | | Structure of Chaetomium thermophilum mApe1 | | Descriptor: | Aminopeptidase-like protein, ZINC ION | | Authors: | Bertipaglia, C, Jakobi, A.J, Wilmanns, M, Sachse, C. | | Deposit date: | 2016-04-28 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.758 Å) | | Cite: | Higher-order assemblies of oligomeric cargo receptor complexes form the membrane scaffold of the Cvt vesicle.
Embo Rep., 17, 2016
|
|
9EOO
 
 | |
9EOM
 
 | |
9EON
 
 | |
9EOP
 
 | |
4UFT
 
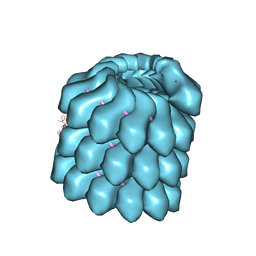 | | Structure of the helical Measles virus nucleocapsid | | Descriptor: | 5'-R(*CP*CP*CP*CP*CP*CP)-3', NUCLEOPROTEIN | | Authors: | Gutsche, I, Desfosses, A, Effantin, G, Ling, W.L, Haupt, M, Ruigrok, R.W.H, Sachse, C, Schoehn, G. | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-Atomic Cryo-Em Structure of the Helical Measles Virus Nucleocapsid.
Science, 348, 2015
|
|
4UF8
 
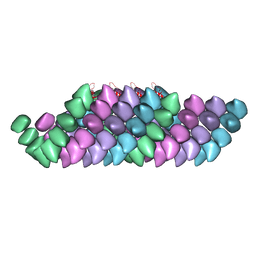 | | Electron cryo-microscopy structure of PB1-p62 filaments | | Descriptor: | SEQUESTOSOME-1 | | Authors: | Ciuffa, R, Lamark, T, Tarafder, A, Guesdon, A, Rybina, S, Hagen, W.J.H, Johansen, T, Sachse, C. | | Deposit date: | 2015-03-15 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.9 Å) | | Cite: | The Selective Autophagy Receptor P62 Forms a Flexible Filamentous Helical Scaffold.
Cell Rep., 11, 2015
|
|
4UDV
 
 | | Cryo-EM structure of TMV at 3.35 A resolution | | Descriptor: | 5'-D(*GP*AP*AP)-3', CAPSID PROTEIN | | Authors: | Fromm, S.A, Bharat, T.A.M, Jakobi, A.J, Hagen, W.J.H, Sachse, C. | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Seeing Tobacco Mosaic Virus Through Direct Electron Detectors.
J.Struct.Biol., 189, 2015
|
|
5KC2
 
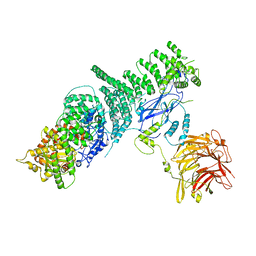 | | Negative stain structure of Vps15/Vps34 complex | | Descriptor: | Phosphatidylinositol 3-kinase VPS34, Serine/threonine-protein kinase VPS15 | | Authors: | Kirsten, M.L, Zhang, L, Ohashi, Y, Perisic, O, Williams, R.L, Sachse, C. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
5AI7
 
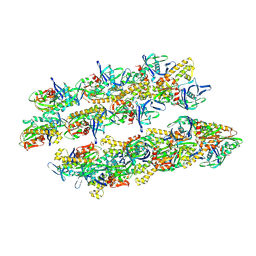 | | ParM doublet model | | Descriptor: | PLASMID SEGREGATION PROTEIN PARM | | Authors: | Bharat, T.A.M, Murshudov, G.N, Sachse, C, Lowe, J. | | Deposit date: | 2015-02-12 | | Release date: | 2015-04-22 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structures of Actin-Like Parm Filaments Show Architecture of Plasmid-Segregating Spindles
Nature, 523, 2015
|
|
5AHV
 
 | | Cryo-EM structure of helical ANTH and ENTH tubules on PI(4,5)P2-containing membranes | | Descriptor: | ANTH DOMAIN OF ENDOCYTIC ADAPTOR SLA2, ENTH DOMAIN OF EPSIN ENT1 | | Authors: | Skruzny, M, Desfosses, A, Prinz, S, Dodonova, S.O, Gieras, A, Uetrecht, C, Jakobi, A.J, Abella, M, Hagen, W.J.H, Schulz, J, Meijers, R, Rybin, V, Briggs, J.A.G, Sachse, C, Kaksonen, M. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | An Organized Co-Assembly of Clathrin Adaptors is Essential for Endocytosis.
Dev.Cell, 33, 2015
|
|
