1GU0
 
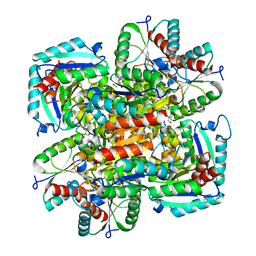 | | CRYSTAL STRUCTURE OF TYPE II DEHYDROQUINASE FROM STREPTOMYCES COELICOLOR | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Roszak, A.W, Krell, T, Robinson, D, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces Coelicolor
Structure, 10, 2002
|
|
1GTZ
 
 | | Structure of STREPTOMYCES COELICOLOR TYPE II DEHYDROQUINASE R23A MUTANT IN COMPLEX WITH DEHYDROSHIKIMATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, 3-DEHYDROSHIKIMATE | | Authors: | Roszak, A.W, Krell, T, Robinson, D.A, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces Coelicolor
Structure, 10, 2002
|
|
1PYH
 
 | | Crystal structure of RC-LH1 core complex from Rhodopseudomonas palustris | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Roszak, A.W, Howard, T.D, Southall, J, Gardiner, A.T, Law, C.J, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2003-07-08 | | Release date: | 2004-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Crystal structure of the RC-LH1 core complex from Rhodopseudomonas palustris.
Science, 302, 2003
|
|
1D0I
 
 | | CRYSTAL STRUCTURE OF TYPE II DEHYDROQUINASE FROM STREPTOMYCES COELICOLOR COMPLEXED WITH PHOSPHATE IONS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, TYPE II 3-DEHYDROQUINATE HYDRATASE | | Authors: | Roszak, A.W, Krell, T, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 1999-09-10 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure and mechanism of the type II dehydroquinase from Streptomyces coelicolor.
Structure, 10, 2002
|
|
1GU1
 
 | | Crystal structure of type II dehydroquinase from Streptomyces coelicolor complexed with 2,3-anhydro-quinic acid | | Descriptor: | 2,3 -ANHYDRO-QUINIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Roszak, A.W, Robinson, D.A, Krell, T, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces Coelicolor
Structure, 10, 2002
|
|
1RGN
 
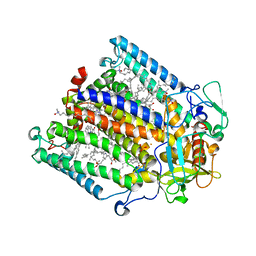 | | Structure of the reaction centre from Rhodobacter sphaeroides carotenoidless strain R-26.1 reconstituted with spheroidene | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Roszak, A.W, Frank, H.A, McKendrick, K, Mitchell, I.A, Cogdell, R.J, Isaacs, N.W. | | Deposit date: | 2003-11-12 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein Regulation of Carotenoid Binding: Gatekeeper and Locking Amino Acid Residues in Reaction Centers of Rhodobacter sphaeroides
STRUCTURE, 12, 2004
|
|
1RG5
 
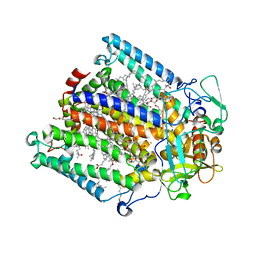 | | Structure of the photosynthetic reaction centre from Rhodobacter sphaeroides carotenoidless strain R-26.1 | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Roszak, A.W, Hashimoto, H, Gardiner, A.T, Cogdell, R.J, Isaacs, N.W. | | Deposit date: | 2003-11-11 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Regulation of Carotenoid Binding: Gatekeeper and Locking Amino Acid Residues in Reaction Centers of Rhodobacter sphaeroides
STRUCTURE, 12, 2004
|
|
1RQK
 
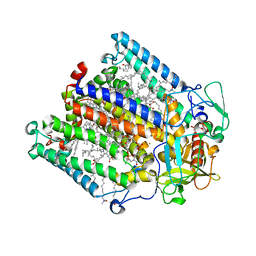 | | Structure of the reaction centre from Rhodobacter sphaeroides carotenoidless strain R-26.1 reconstituted with 3,4-dihydrospheroidene | | Descriptor: | 3,4-DIHYDROSPHEROIDENE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Roszak, A.W, Hashimoto, H, Gardiner, A.T, Cogdell, R.J, Isaacs, N.W. | | Deposit date: | 2003-12-05 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein Regulation of Carotenoid Binding: Gatekeeper and Locking Amino Acid Residues in Reaction Centers of Rhodobacter sphaeroides
STRUCTURE, 12, 2004
|
|
2HG9
 
 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with tetrabrominated phosphatidylcholine | | Descriptor: | (7R,18S,19R)-18,19-DIBROMO-7-{[(9S,10S)-9,10-DIBROMOOCTADECANOYL]OXY}-4-HYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-P HOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-26 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
2HH1
 
 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with dibrominated phosphatidylcholine | | Descriptor: | (7R,14S)-14,15-DIBROMO-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-27 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
2HJ6
 
 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with dibrominated phosphatidylserine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-30 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
2HIT
 
 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with dibrominated phosphatidylethanolamine | | Descriptor: | (1R)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (9S,10S)-9,10-DIBROMOOCTADECANOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-29 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
2HHK
 
 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with dibrominated phosphatidylglycerol | | Descriptor: | (1R)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (9S,10S)-9,10-DIBROMOOCTADECANOATE, (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-28 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
2HG3
 
 | | Reaction centre from Rhodobacter sphaeroides strain R-26.1 complexed with brominated phosphatidylcholine | | Descriptor: | (7R,14S)-14,15-DIBROMO-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2006-06-26 | | Release date: | 2007-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Brominated Lipids Identify Lipid Binding Sites on the Surface of the Reaction Center from Rhodobacter sphaeroides.
Biochemistry, 46, 2007
|
|
3T6E
 
 | | Crystal Structure of the Reaction Centre from Blastochloris viridis strain DSM 133 (ATCC 19567) substrain-94 | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | New insights into the structure of the reaction centre from Blastochloris viridis: evolution in the laboratory.
Biochem.J., 442, 2012
|
|
3T6D
 
 | | Crystal Structure of the Reaction Centre from Blastochloris viridis strain DSM 133 (ATCC 19567) substrain-08 | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New insights into the structure of the reaction centre from Blastochloris viridis: evolution in the laboratory.
Biochem.J., 442, 2012
|
|
1H0R
 
 | | Type II Dehydroquinase from Mycobacterium tuberculosis complexed with 2,3-anhydro-quinic acid | | Descriptor: | 2,3 -ANHYDRO-QUINIC ACID, 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | Roszak, A.W, Robinson, D.A, Frederickson, M, Abell, C, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-06-27 | | Release date: | 2003-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Selectivity of Oxime Based Inhibitors Towards Type II Dehydroquinase from Mycobacterium Tuberculosis
To be Published
|
|
1NYT
 
 | | SHIKIMATE DEHYDROGENASE AroE COMPLEXED WITH NADP+ | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Roszak, A.W, Lapthorn, A.J. | | Deposit date: | 2003-02-13 | | Release date: | 2003-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of shikimate dehydrogenase AroE and its Paralog YdiB. A common structural framework for different activities.
J.Biol.Chem., 278, 2003
|
|
4FZL
 
 | | High resolution structure of truncated bacteriocin syringacin M from Pseudomonas syringae pv. tomato DC3000 | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin, CALCIUM ION, ... | | Authors: | Roszak, A.W, Grinter, R, Cogdell, J.R, Walker, D. | | Deposit date: | 2012-07-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The Crystal Structure of the Lipid II-degrading Bacteriocin Syringacin M Suggests Unexpected Evolutionary Relationships between Colicin M-like Bacteriocins.
J.Biol.Chem., 287, 2012
|
|
4FZN
 
 | |
4FZM
 
 | | Crystal structure of the bacteriocin syringacin M from Pseudomonas syringae pv. tomato DC3000 | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin, CALCIUM ION | | Authors: | Roszak, A.W, Grinter, R, Cogdell, J.R, Walker, D. | | Deposit date: | 2012-07-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | The Crystal Structure of the Lipid II-degrading Bacteriocin Syringacin M Suggests Unexpected Evolutionary Relationships between Colicin M-like Bacteriocins.
J.Biol.Chem., 287, 2012
|
|
1H05
 
 | |
1H0S
 
 | | 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with 3-hydroxyimino-quinic acid | | Descriptor: | 3-DEHYDROQUINATE DEHYDRATASE, 3-HYDROXYIMINO QUINIC ACID, GLYCEROL, ... | | Authors: | Roszak, A.W, Frederickson, M, Abell, C, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-06-27 | | Release date: | 2003-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Specificity of Oxime Based Inhibitors Towards Type II Dehydroquinase from Mycobacterium Tuberculosis
To be Published
|
|
1V1J
 
 | | Crystal structure of type II Dehydroquintae Dehydratase from Streptomyces coelicolor in complex with 3-fluoro | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-ANHYDRO-3-FLUORO-QUINIC ACID, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Roszak, A.W, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2004-04-16 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | (1R,4S,5R)-3-Fluoro-1,4,5-Trihydroxy-2-Cyclohexene-1-Carboxylic Acid: The Fluoro Analogue of the Enolate Intermediate in the Reaction Catalyzed by Type II Dehydroquinases
Org.Biomol.Chem., 2, 2004
|
|
4LEA
 
 | | The Crystal Structure of Pyocin L1 bound to D-mannose at 2.55 Angstroms | | Descriptor: | Pyocin L1, beta-D-mannopyranose | | Authors: | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, C.J, Walker, D. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
