1NBA
 
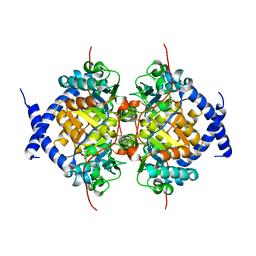 | | CRYSTAL STRUCTURE ANALYSIS, REFINEMENT AND ENZYMATIC REACTION MECHANISM OF N-CARBAMOYLSARCOSINE AMIDOHYDROLASE FROM ARTHROBACTER SP. AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | N-CARBAMOYLSARCOSINE AMIDOHYDROLASE, SULFATE ION | | Authors: | Romao, M.J, Turk, D, Gomis-Ruth, F.-Z, Huber, R, Schumacher, G, Mollering, H, Russmann, L. | | Deposit date: | 1992-05-18 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis, refinement and enzymatic reaction mechanism of N-carbamoylsarcosine amidohydrolase from Arthrobacter sp. at 2.0 A resolution.
J.Mol.Biol., 226, 1992
|
|
1SFP
 
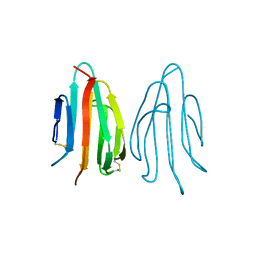 | | CRYSTAL STRUCTURE OF ACIDIC SEMINAL FLUID PROTEIN (ASFP) AT 1.9 A RESOLUTION: A BOVINE POLYPEPTIDE FROM THE SPERMADHESIN FAMILY | | Descriptor: | ASFP | | Authors: | Romao, M.J, Kolln, I, Dias, J.M, Carvalho, A.L, Romero, A, Varela, P.F, Sanz, L, Topfer-Petersen, E, Calvete, J.J. | | Deposit date: | 1997-06-24 | | Release date: | 1998-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of two spermadhesins reveal the CUB domain fold.
Nat.Struct.Biol., 4, 1997
|
|
6Q6Q
 
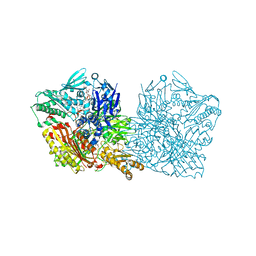 | | Human aldehyde oxidase SNP G1269R | | Descriptor: | Aldehyde oxidase, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mota, C, Coelho, C, Santos-Silva, T, Romao, M.J. | | Deposit date: | 2018-12-11 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.10003781 Å) | | Cite: | Human aldehyde oxidase (hAOX1): structure determination of the Moco-free form of the natural variant G1269R and biophysical studies of single nucleotide polymorphisms.
Febs Open Bio, 9, 2019
|
|
4UWV
 
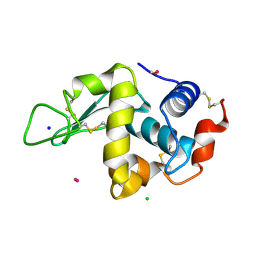 | | Lysozyme soaked with a ruthenium based CORM with a pyridine ligand (complex 8) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, LYSOZYME C, ... | | Authors: | Santos, M.F.A, Mukhopadhyay, A, Romao, M.J, Romao, C.C, Santos-Silva, T. | | Deposit date: | 2014-08-14 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A Contribution to the Rational Design of Ru(Co)3Cl2L Complexes for in Vivo Delivery of Co.
Dalton Trans, 44, 2015
|
|
4UWU
 
 | | Lysozyme soaked with a ruthenium based CORM with a pyridine ligand (complex 7) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Santos, M.F.A, Mukhopadhyay, A, Romao, M.J, Romao, C.C, Santos-Silva, T. | | Deposit date: | 2014-08-14 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Contribution to the Rational Design of Ru(Co)3Cl2L Complexes for in Vivo Delivery of Co.
Dalton Trans, 44, 2015
|
|
4UWN
 
 | | Lysozyme soaked with a ruthenium based CORM with a methione oxide ligand (complex 6b) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Santos, M.F.A, Mukhopadhyay, A, Romao, M.J, Romao, C.C, Santos-Silva, T. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A Contribution to the Rational Design of Ru(Co)3Cl2L Complexes for in Vivo Delivery of Co.
Dalton Trans, 44, 2015
|
|
6YIR
 
 | |
1VLB
 
 | | STRUCTURE REFINEMENT OF THE ALDEHYDE OXIDOREDUCTASE FROM DESULFOVIBRIO GIGAS AT 1.28 A | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, CHLORIDE ION, ... | | Authors: | Rebelo, J.M, Dias, J.M, Huber, R, Moura, J.J.G, Romao, M.J. | | Deposit date: | 2004-07-20 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure refinement of the aldehyde oxidoreductase from Desulfovibrio gigas (MOP) at 1.28 A
J.Biol.Inorg.Chem., 6, 2001
|
|
4US8
 
 | | Aldehyde Oxidoreductase from Desulfovibrio gigas (MOP), soaked with benzaldehyde | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, BICARBONATE ION, ... | | Authors: | Correia, H.D, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Aromatic Aldehydes at the Active Site of Aldehyde Oxidoreductase from Desulfovibrio Gigas: Reactivity and Molecular Details of the Enzyme-Substrate and Enzyme-Product Interaction.
J.Biol.Inorg.Chem., 20, 2015
|
|
4USA
 
 | | Aldehyde Oxidoreductase from Desulfovibrio gigas (MOP), soaked with trans-cinnamaldehyde | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, BICARBONATE ION, ... | | Authors: | Correia, H.D, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Aromatic Aldehydes at the Active Site of Aldehyde Oxidoreductase from Desulfovibrio Gigas: Reactivity and Molecular Details of the Enzyme-Substrate and Enzyme-Product Interaction.
J.Biol.Inorg.Chem., 20, 2015
|
|
4US9
 
 | | Aldehyde Oxidoreductase from Desulfovibrio gigas (MOP), soaked with 3- phenylpropionaldehyde | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), 3-PHENYLPROPANAL, ALDEHYDE OXIDOREDUCTASE, ... | | Authors: | Correia, H.D, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Aromatic Aldehydes at the Active Site of Aldehyde Oxidoreductase from Desulfovibrio Gigas: Reactivity and Molecular Details of the Enzyme-Substrate and Enzyme-Product Interaction.
J.Biol.Inorg.Chem., 20, 2015
|
|
4UHW
 
 | | Human aldehyde oxidase | | Descriptor: | ALDEHYDE OXIDASE, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Coelho, C, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2015-03-26 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights Into Xenobiotic and Inhibitor Binding to Human Aldehyde Oxidase
Nat.Chem.Biol., 11, 2015
|
|
4UHX
 
 | | Human aldehyde oxidase in complex with phthalazine and thioridazine | | Descriptor: | 10-{2-[(2R)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, 10-{2-[(2S)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, ALDEHYDE OXIDASE, ... | | Authors: | Coelho, C, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2015-03-26 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights Into Xenobiotic and Inhibitor Binding to Human Aldehyde Oxidase
Nat.Chem.Biol., 11, 2015
|
|
7Z5O
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Dithionite reduced form | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, Formate dehydrogenase, ... | | Authors: | Mota, C, Oliveira, A.R, Klymanska, K, Pereira, I.C, Romao, M.J. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.527 Å) | | Cite: | Spectroscopic and Structural Characterization of Reduced Desulfovibrio vulgaris Hildenborough W-FdhAB Reveals Stable Metal Coordination during Catalysis.
Acs Chem.Biol., 17, 2022
|
|
7ZS8
 
 | | Mixed-valence, active form, of cytochrome c peroxidase from obligate human pathogenic bacterium Neisseria gonorrhoeae at 1.4 Angstrom resolution | | Descriptor: | CALCIUM ION, Cytochrome-c peroxidase, HEME C, ... | | Authors: | Carvalho, A.L, Romao, M.J, Pauleta, S, Nobrega, C. | | Deposit date: | 2022-05-06 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Characterization of Neisseria gonorrhoeae Bacterial Peroxidase-Insights into the Catalytic Cycle of Bacterial Peroxidases.
Int J Mol Sci, 24, 2023
|
|
3ML1
 
 | | Crystal Structure of the Periplasmic Nitrate Reductase from Cupriavidus necator | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DIOXOTHIOMOLYBDENUM(VI) ION, Diheme cytochrome c napB, ... | | Authors: | Coelho, C, Trincao, J, Romao, M.J. | | Deposit date: | 2010-04-16 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Cupriavidus necator nitrate reductase in oxidized and partially reduced states
J.Mol.Biol., 408, 2011
|
|
3O5A
 
 | | Crystal Structure of partially reduced Periplasmic Nitrate Reductase from Cupriavidus necator using Ionic Liquids | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CHLORIDE ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Coelho, C, Trincao, J, Romao, M.J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The crystal structure of Cupriavidus necator nitrate reductase in oxidized and partially reduced states
J.Mol.Biol., 408, 2011
|
|
7ORC
 
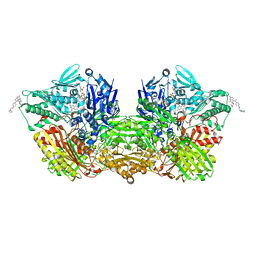 | | Human Aldehyde Oxidase in complex with Raloxifene | | Descriptor: | Aldehyde oxidase, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Mota, C, Coelho, C, Santos Silva, T, Romao, M.J. | | Deposit date: | 2021-06-05 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interrogating the Inhibition Mechanisms of Human Aldehyde Oxidase by X-ray Crystallography and NMR Spectroscopy: The Raloxifene Case.
J.Med.Chem., 64, 2021
|
|
7OPN
 
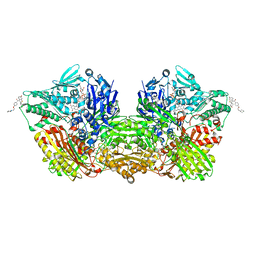 | | Human Aldehyde Oxidase SNP R1231H in complex with Raloxifene | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Aldehyde oxidase, DIMETHYL SULFOXIDE, ... | | Authors: | Mota, C, Coelho, C, Santos Silva, T, Romao, M.J. | | Deposit date: | 2021-06-01 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interrogating the Inhibition Mechanisms of Human Aldehyde Oxidase by X-ray Crystallography and NMR Spectroscopy: The Raloxifene Case.
J.Med.Chem., 64, 2021
|
|
6QKN
 
 | | Structure of the azide-inhibited form of cytochrome c peroxidase from obligate human pathogenic bacterium Neisseria gonorrhoeae | | Descriptor: | AZIDE ION, CALCIUM ION, Cytochrome-c peroxidase, ... | | Authors: | Carvalho, A.L, Romao, M.J, Pauleta, S, Nobrega, C. | | Deposit date: | 2019-01-29 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the mixed-valence, active form, of cytochrome c peroxidase from obligate human pathogenic bacterium Neisseria gonorrhoeae
To Be Published
|
|
3ERX
 
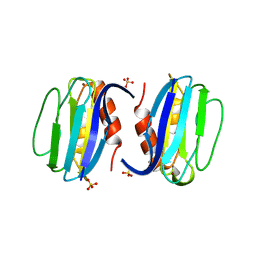 | | High-resolution structure of Paracoccus pantotrophus pseudoazurin | | Descriptor: | COPPER (II) ION, Pseudoazurin, SULFATE ION | | Authors: | Najmudin, S, Pauleta, S.R, Moura, I, Romao, M.J. | | Deposit date: | 2008-10-03 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The 1.4 A resolution structure of Paracoccus pantotrophus pseudoazurin.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3FAH
 
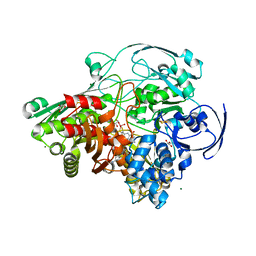 | | Glycerol inhibited form of Aldehyde oxidoreductase from Desulfovibrio gigas | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), Aldehyde oxidoreductase, CHLORIDE ION, ... | | Authors: | Santos-Silva, T, Romao, M.J. | | Deposit date: | 2008-11-17 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Kinetic, structural, and EPR studies reveal that aldehyde oxidoreductase from Desulfovibrio gigas does not need a sulfido ligand for catalysis and give evidence for a direct Mo-C interaction in a biological system.
J.Am.Chem.Soc., 131, 2009
|
|
3FC4
 
 | | Ethylene glycol inhibited form of Aldehyde oxidoreductase from Desulfovibrio gigas | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), 1,2-ETHANEDIOL, Aldehyde oxidoreductase, ... | | Authors: | Santos-Silva, T, Romao, M.J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Kinetic, structural, and EPR studies reveal that aldehyde oxidoreductase from Desulfovibrio gigas does not need a sulfido ligand for catalysis and give evidence for a direct Mo-C interaction in a biological system.
J.Am.Chem.Soc., 131, 2009
|
|
5G5H
 
 | | Escherichia coli Periplasmic Aldehyde Oxidase R440H mutant | | Descriptor: | ACETATE ION, Aldehyde oxidoreductase FAD-binding subunit PaoB, Aldehyde oxidoreductase iron-sulfur-binding subunit PaoA, ... | | Authors: | Correia, M.A.S, Otrelo-Cardoso, A.R, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Escherichia Coli Periplasmic Aldehyde Oxidoreductase is an Exceptional Member of the Xanthine Oxidase Family of Molybdoenzymes.
Acs Chem.Biol., 11, 2016
|
|
5G5G
 
 | | Escherichia coli Periplasmic Aldehyde Oxidase | | Descriptor: | ACETATE ION, CHLORIDE ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Correia, M.A.S, Otrelo-Cardoso, A.R, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Escherichia Coli Periplasmic Aldehyde Oxidoreductase is an Exceptional Member of the Xanthine Oxidase Family of Molybdoenzymes.
Acs Chem.Biol., 11, 2016
|
|
