1PM6
 
 | | Solution Structure of Full-Length Excisionase (Xis) from Bacteriophage HK022 | | Descriptor: | Excisionase | | Authors: | Rogov, V.V, Luecke, C, Muresanu, L, Wienk, H, Kleinhaus, I, Werner, K, Loehr, F, Pristovsek, P, Rueterjans, H. | | Deposit date: | 2003-06-10 | | Release date: | 2003-12-30 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability of the full-length excisionase from bacteriophage HK022.
Eur.J.Biochem., 270, 2003
|
|
1SR2
 
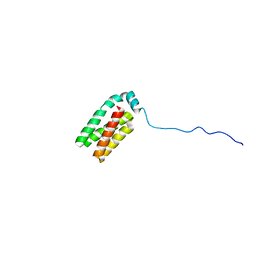 | |
2AYY
 
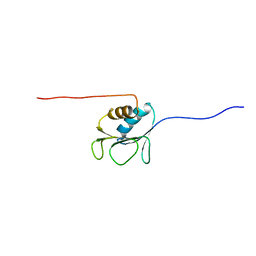 | | Solution structure of the E.coli RcsC C-terminus (residues 700-816) containing linker region | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
2AYZ
 
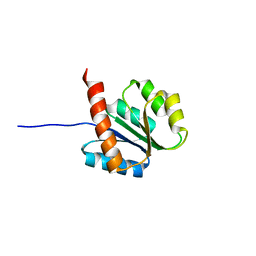 | | Solution structure of the E.coli RcsC C-terminus (residues 817-949) containing phosphoreceiver domain | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
2AYX
 
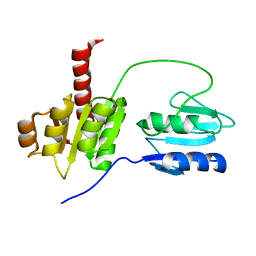 | | Solution structure of the E.coli RcsC C-terminus (residues 700-949) containing linker region and phosphoreceiver domain | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
2L8Y
 
 | | Solution structure of the E. coli outer membrane protein RcsF (periplasmatic domain) | | Descriptor: | Protein rcsF | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Lohr, F, Doetsch, V. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A disulfide bridge network within the soluble periplasmic domain determines structure and function of the outer membrane protein RCSF.
J.Biol.Chem., 286, 2011
|
|
2LUE
 
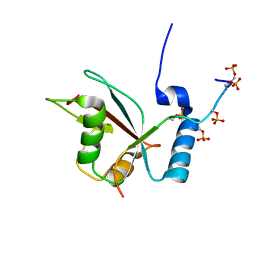 | | LC3B OPTN-LIR Ptot complex structure | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, Optineurin | | Authors: | Rogov, V.V, Rozenknop, A, Loehr, F, Guentert, P, Doetsch, V. | | Deposit date: | 2012-06-13 | | Release date: | 2013-07-17 | | Last modified: | 2022-08-24 | | Method: | SOLUTION NMR | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella.
Biochem.J., 454, 2013
|
|
2L8J
 
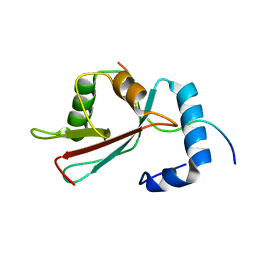 | | GABARAPL-1 NBR1-LIR complex structure | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, NBR1-LIR peptide | | Authors: | Rogov, V.V, Rozenknop, A, Rogova, N.Y, Loehr, F, Guentert, P, Dikic, I, Doetsch, V. | | Deposit date: | 2011-01-17 | | Release date: | 2011-05-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Characterization of the Interaction of GABARAPL-1 with the LIR Motif of NBR1.
J.Mol.Biol., 410, 2011
|
|
2KX7
 
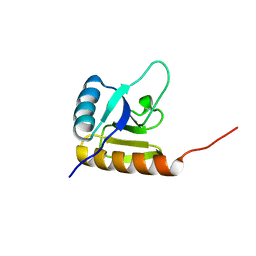 | | Solution structure of the E.coli RcsD-ABL domain (residues 688-795) | | Descriptor: | Sensor-like histidine kinase yojN | | Authors: | Rogov, V.V, Schmoee, K, Rogova, N.Y, Loehr, F, Bernhard, F, Doetsch, V. | | Deposit date: | 2010-04-27 | | Release date: | 2011-04-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into Rcs Phosphotransfer: The Newly Identified RcsD-ABL Domain Enhances Interaction with the Response Regulator RcsB.
Structure, 19, 2011
|
|
5HKH
 
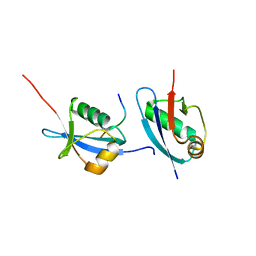 | | Crystal structure of Ufm1 in complex with UBA5 | | Descriptor: | ASP-ASN-GLU-TRP-GLY-ILE-GLU-LEU-VAL, Ubiquitin-fold modifier 1 | | Authors: | Huber, J, Doetsch, V, Rogov, V.V, Akutsu, M. | | Deposit date: | 2016-01-14 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Functional Analysis of a Novel Interaction Motif within UFM1-activating Enzyme 5 (UBA5) Required for Binding to Ubiquitin-like Proteins and Ufmylation.
J.Biol.Chem., 291, 2016
|
|
6H8C
 
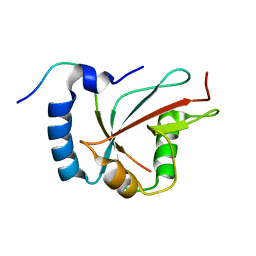 | | Structure of the human GABARAPL2 protein in complex with the UBA5 LIR motif | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 2, Ubiquitin-like modifier-activating enzyme 5 | | Authors: | Huber, J, Loehr, F, Gruber, J, Akutsu, M, Guentert, P, Doetsch, V, Rogov, V.V. | | Deposit date: | 2018-08-02 | | Release date: | 2019-05-01 | | Last modified: | 2020-01-29 | | Method: | SOLUTION NMR | | Cite: | An atypical LIR motif within UBA5 (ubiquitin like modifier activating enzyme 5) interacts with GABARAP proteins and mediates membrane localization of UBA5.
Autophagy, 16, 2020
|
|
7OVC
 
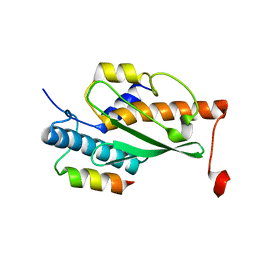 | | Structure of the human UFC1 protein in complex with the UBA5 C-terminal UFC1-binding motif. | | Descriptor: | Ubiquitin-fold modifier-conjugating enzyme 1, Ubiquitin-like modifier-activating enzyme 5 | | Authors: | Wesch, W, Loehr, F, Rogova, N, Doetsch, V, Rogov, V.V. | | Deposit date: | 2021-06-14 | | Release date: | 2021-08-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Concerted Action of UBA5 C-Terminal Unstructured Regions Is Important for Transfer of Activated UFM1 to UFC1.
Int J Mol Sci, 22, 2021
|
|
1I6E
 
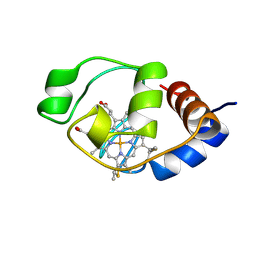 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE OXIDIZED STATE | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Reincke, B, Perez, C, Pristovsek, P, Luecke, C, Ludwig, C, Loehr, F, Rogov, V.V, Ludwig, B, Rueterjans, H. | | Deposit date: | 2001-03-02 | | Release date: | 2001-10-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the functional domain of Paracoccus denitrificans cytochrome c(552) in both redox states.
Biochemistry, 40, 2001
|
|
1I6D
 
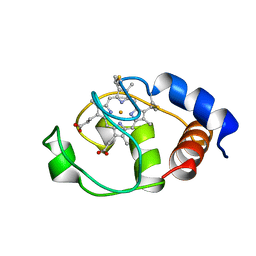 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE REDUCED STATE | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Reincke, B, Perez, C, Pristovsek, P, Luecke, C, Ludwig, C, Loehr, F, Rogov, V.V, Ludwig, B, Rueterjans, H. | | Deposit date: | 2001-03-02 | | Release date: | 2001-10-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the functional domain of Paracoccus denitrificans cytochrome c(552) in both redox states.
Biochemistry, 40, 2001
|
|
5DCN
 
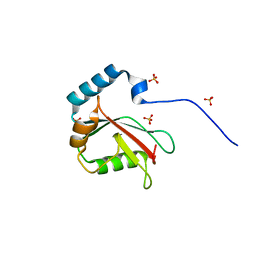 | | Crystal structure of LC3 in complex with TECPR2 LIR | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Stadel, D, Huber, J, Dotsch, V, Rogov, V.V, Behrends, C, Akutsu, M. | | Deposit date: | 2015-08-24 | | Release date: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TECPR2 Cooperates with LC3C to Regulate COPII-Dependent ER Export.
Mol.Cell, 60, 2015
|
|
2MYX
 
 | | Structure of the CUE domain of yeast Cue1 | | Descriptor: | Coupling of ubiquitin conjugation to ER degradation protein 1 | | Authors: | Kniss, A, Rogov, V.V, Loehr, F, Guentert, P, Doetsch, V. | | Deposit date: | 2015-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The CUE Domain of Cue1 Aligns Growing Ubiquitin Chains with Ubc7 for Rapid Elongation.
Mol.Cell, 62, 2016
|
|
2RON
 
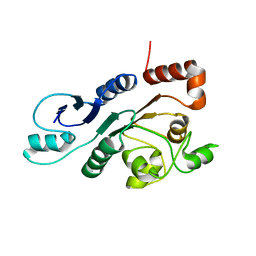 | | The external thioesterase of the Surfactin-Synthetase | | Descriptor: | Surfactin synthetase thioesterase subunit | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guentert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-12 | | Last modified: | 2016-10-26 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase
Nature, 454, 2008
|
|
2GDX
 
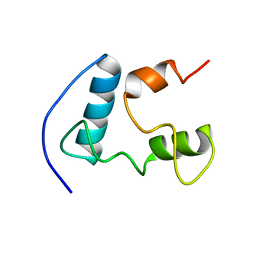 | | Solution structure of the B. brevis TycC3-PCP in H-state | | Descriptor: | Tyrocidine synthetase III | | Authors: | Koglin, A, Loehr, F, Rogov, V.V, Marahiel, M.A, Bernhard, F, Doetsch, V. | | Deposit date: | 2006-03-17 | | Release date: | 2006-08-01 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Conformational switches modulate protein interactions in peptide antibiotic synthetases
Science, 312, 2006
|
|
2GDW
 
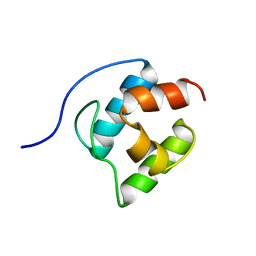 | | Solution structure of the B. brevis TycC3-PCP in A/H-state | | Descriptor: | Tyrocidine synthetase III | | Authors: | Koglin, A, Loehr, F, Rogov, V.V, Marahiel, M.A, Bernhard, F, Doetsch, V. | | Deposit date: | 2006-03-17 | | Release date: | 2006-08-01 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Conformational switches modulate protein interactions in peptide antibiotic synthetases
Science, 312, 2006
|
|
2GDY
 
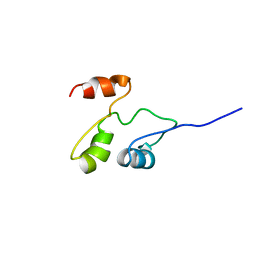 | | Solution structure of the B. brevis TycC3-PCP in A-state | | Descriptor: | Tyrocidine synthetase III | | Authors: | Koglin, A, Loehr, F, Rogov, V.V, Marahiel, M.A, Bernhard, F, Doetsch, V. | | Deposit date: | 2006-03-17 | | Release date: | 2006-08-01 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Conformational switches modulate protein interactions in peptide antibiotic synthetases
Science, 312, 2006
|
|
2K2Q
 
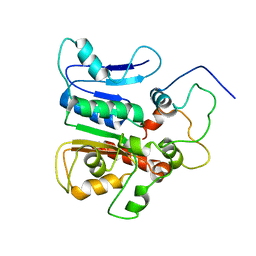 | | complex structure of the external thioesterase of the Surfactin-synthetase with a carrier domain | | Descriptor: | Surfactin synthetase thioesterase subunit, Tyrocidine synthetase 3 (Tyrocidine synthetase III) | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guntert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Dotsch, V. | | Deposit date: | 2008-04-10 | | Release date: | 2008-12-09 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase.
Nature, 454, 2008
|
|
4WAA
 
 | | Crystal structure of Nix LIR-fused human LC3B_2-119 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Suzuki, H, Ravichandran, A.C, Dobson, R.C.J, Novak, I, Wakatsuki, S. | | Deposit date: | 2014-08-29 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins.
Sci Rep, 7, 2017
|
|
3VTW
 
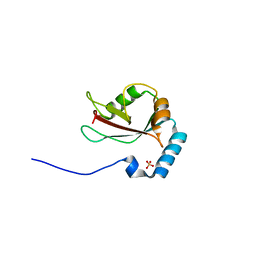 | | Crystal structure of T7-tagged Optineurin LIR-fused human LC3B_2-119 | | Descriptor: | Optineurin, microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-06-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
3VTU
 
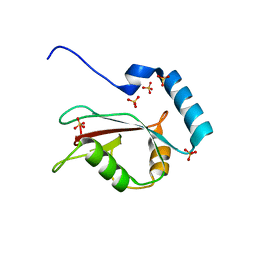 | | Crystal structure of human LC3B_2-119 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-06-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
3VTV
 
 | | Crystal structure of Optineurin LIR-fused human LC3B_2-119 | | Descriptor: | Optineurin, microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-06-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
