1HL4
 
 | | The Structure of Apo Type Human Cu, Zn Superoxide Dismutase | | Descriptor: | SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Strange, R.W, Antonyuk, S, Hough, M.A, Doucette, P, Rodriguez, J, Hart, P.J, Hayward, L.J, Valentine, J.S, Hasnain, S.S. | | Deposit date: | 2003-03-13 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Structure of Holo and Metal-Deficient Wild-Type Human Cu, Zn Superoxide Dismutase and its Relevance to Familial Amyotrophic Lateral Sclerosis
J.Mol.Biol., 328, 2003
|
|
1HL5
 
 | | The Structure of Holo Type Human Cu, Zn Superoxide Dismutase | | Descriptor: | CALCIUM ION, COPPER (II) ION, SUPEROXIDE DISMUTASE, ... | | Authors: | Strange, R.W, Antonyuk, S, Hough, M.A, Doucette, P, Rodriguez, J, Hart, P.J, Hayward, L.J, Valentine, J.S, Hasnain, S.S. | | Deposit date: | 2003-03-13 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Holo and Metal-Deficient Wild-Type Human Cu, Zn Superoxide Dismutase and its Relevance to Familial Amyotrophic Lateral Sclerosis
J.Mol.Biol., 328, 2003
|
|
1S4B
 
 | | Crystal structure of human thimet oligopeptidase. | | Descriptor: | Thimet oligopeptidase, ZINC ION | | Authors: | Ray, K, Hines, C.S, Coll-Rodriguez, J, Rodgers, D.W. | | Deposit date: | 2004-01-15 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human thimet oligopeptidase provides insight into substrate recognition, regulation, and localization
J.Biol.Chem., 279, 2004
|
|
4QX0
 
 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QX3
 
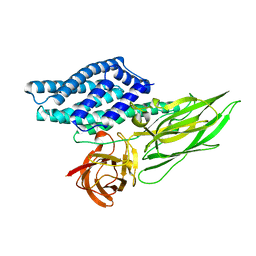 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QX2
 
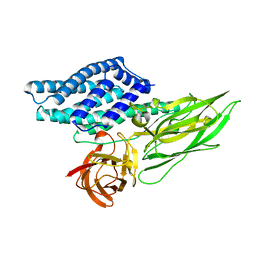 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QX1
 
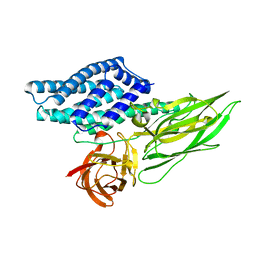 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1OEZ
 
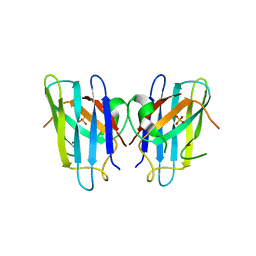 | | Zn His46Arg mutant of Human Cu, Zn Superoxide Dismutase | | Descriptor: | SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ZINC ION | | Authors: | Strange, R.W, Antonyuk, S, Hough, M.A, Doucette, P, Rodriguez, J, Elam, J.S, Hart, P.J, Hayward, L.J, Valentine, J.S, Hasnain, S.S. | | Deposit date: | 2003-04-02 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Amyloid-Like Filaments and Water-Filled Nanotubes Formed by Sod1 Mutant Proteins Linked to Familial Als
Nat.Struct.Biol., 10, 2003
|
|
5K7R
 
 | | MicroED structure of trypsin at 1.7 A resolution | | Descriptor: | CALCIUM ION, Cationic trypsin | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.7 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7T
 
 | | MicroED structure of thermolysin at 2.5 A resolution | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7Q
 
 | | MicroED structure of thaumatin at 2.5 A resolution | | Descriptor: | Thaumatin-1 | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7N
 
 | | MicroED structure of tau VQIVYK peptide at 1.1 A resolution | | Descriptor: | VQIVYK | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7S
 
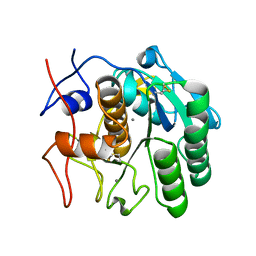 | | MicroED structure of proteinase K at 1.6 A resolution | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.6 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7P
 
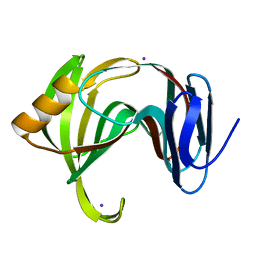 | | MicroED structure of xylanase at 2.3 A resolution | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.3 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7O
 
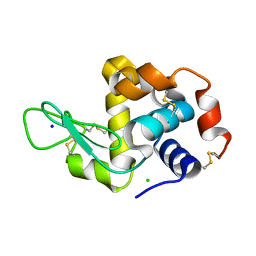 | | MicroED structure of lysozyme at 1.8 A resolution | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.8 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
7NZ8
 
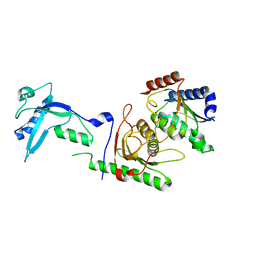 | | Crystal structure of mouse ADAT2/ADAT3 tRNA deamination complex 2 | | Descriptor: | Probable inactive tRNA-specific adenosine deaminase-like protein 3, ZINC ION, tRNA-specific adenosine deaminase 2 | | Authors: | Ramos Morales, E, Romier, C. | | Deposit date: | 2021-03-23 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The structure of the mouse ADAT2/ADAT3 complex reveals the molecular basis for mammalian tRNA wobble adenosine-to-inosine deamination.
Nucleic Acids Res., 49, 2021
|
|
6I9J
 
 | | Human transforming growth factor beta2 in a tetragonal crystal form | | Descriptor: | Transforming growth factor beta-2 proprotein | | Authors: | Gomis-Ruth, F.X, Marino-Puertas, L, del Amo-Maestro, L, Goulas, T. | | Deposit date: | 2018-11-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant production, purification, crystallization, and structure analysis of human transforming growth factor beta 2 in a new conformation.
Sci Rep, 9, 2019
|
|
7QYD
 
 | | mosquitocidal Cry11Ba determined at pH 6.5 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Ba | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P, Sawaya, M.R, Schibrowsky, N.A, Cascio, D, Rodriguez, J.A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7R1E
 
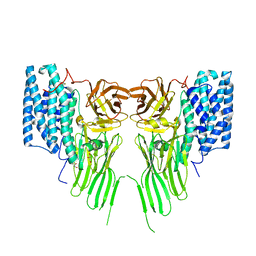 | | Mosquitocidal Cry11Ba determined at pH 10.4 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | GLYCEROL, Pesticidal crystal protein Cry11Ba | | Authors: | Colletier, J.-P, Sawaya, M.R, Schibrowsky, N.A, Cascio, D, Rodriguez, J.A. | | Deposit date: | 2022-02-02 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
2O36
 
 | |
5K2H
 
 | |
5K2F
 
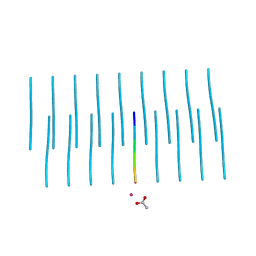 | | Structure of NNQQNY from yeast prion Sup35 with cadmium acetate determined by MicroED | | Descriptor: | ACETATE ION, CADMIUM ION, Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K2E
 
 | | Structure of NNQQNY from yeast prion Sup35 with zinc acetate determined by MicroED | | Descriptor: | ACETIC ACID, Eukaryotic peptide chain release factor GTP-binding subunit, ZINC ION | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K2G
 
 | |
7L7D
 
 | |
