1OB9
 
 | | Holliday Junction Resolving Enzyme | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, HOLLIDAY JUNCTION RESOLVASE | | Authors: | Middleton, C.L, Parker, J.L, Richard, D.J, White, M.F, Bond, C.S. | | Deposit date: | 2003-01-28 | | Release date: | 2004-10-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Recognition and Catalysis by the Holliday Junction Resolving Enzyme Hje.
Nucleic Acids Res., 32, 2004
|
|
1OB8
 
 | | Holliday Junction Resolving Enzyme | | Descriptor: | 1,2-ETHANEDIOL, HOLLIDAY-JUNCTION RESOLVASE, SULFATE ION | | Authors: | Middleton, C.L, Parker, J.L, Richard, D.J, White, M.F, Bond, C.S. | | Deposit date: | 2003-01-28 | | Release date: | 2004-10-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate Recognition and Catalysis by the Holliday Junction Resolving Enzyme Hje.
Nucleic Acids Res., 32, 2004
|
|
4TQX
 
 | | Molecular Basis of Streptococcus mutans Sortase A Inhibition by Chalcone. | | Descriptor: | ACETIC ACID, SULFATE ION, Sortase, ... | | Authors: | Wallock-Richards, D.J, Marles-Wright, J, Clarke, D.J, Maitra, A, Dodds, M, Hanley, B, Campopiano, D.J. | | Deposit date: | 2014-06-12 | | Release date: | 2015-05-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Molecular basis of Streptococcus mutans sortase A inhibition by the flavonoid natural product trans-chalcone.
Chem.Commun.(Camb.), 51, 2015
|
|
2MNA
 
 | | The structural basis of DNA binding by the single-stranded DNA-binding protein from Sulfolobus solfataricus | | Descriptor: | Single-stranded DNA binding protein (SSB), ssDNA | | Authors: | Gamsjaeger, R, Kariawasam, R, Gimenez, A.X, Touma, C.F, McIlwain, E, Bernardo, R.E, Shepherd, N.E, Ataide, S.F, Dong, A.Q, Richard, D.J, White, M.F, Cubeddu, L. | | Deposit date: | 2014-04-02 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structural basis of DNA binding by the single-stranded DNA-binding protein from Sulfolobus solfataricus
Biochem.J., 465, 2015
|
|
3TOR
 
 | | Crystal structure of Escherichia coli NrfA with Europium bound | | Descriptor: | CALCIUM ION, Cytochrome c nitrite reductase, EUROPIUM ION, ... | | Authors: | Lockwood, C.W.J, Clarke, T.A, Butt, J.N, Hemmings, A.M, Richardson, D.J. | | Deposit date: | 2011-09-06 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the active site and calcium binding in cytochrome c nitrite reductases.
Biochem.Soc.Trans., 39, 2011
|
|
3PMQ
 
 | |
2RDZ
 
 | | High Resolution Crystal Structure of the Escherichia coli Cytochrome c Nitrite Reductase. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Hemmings, A.M, RIchardson, D.J. | | Deposit date: | 2007-09-25 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Role of a Conserved Glutamine Residue in Tuning the Catalytic Activity of Escherichia coli Cytochrome c Nitrite Reductase.
Biochemistry, 47, 2008
|
|
2RF7
 
 | | Crystal structure of the escherichia coli nrfa mutant Q263E | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Richardson, D.J, Hemmings, A.M. | | Deposit date: | 2007-09-28 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Role of a Conserved Glutamine Residue in Tuning the Catalytic Activity of Escherichia coli Cytochrome c Nitrite Reductase.
Biochemistry, 47, 2008
|
|
3DAS
 
 | | Structure of the PQQ-bound form of Aldose Sugar Dehydrogenase (Adh) from Streptomyces coelicolor | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Southall, S.M, Doel, J.J, Oubrie, A, Richardson, D.J. | | Deposit date: | 2008-05-30 | | Release date: | 2009-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Enzymatic Characterization of a Thermostable PQQ-dependent Soluble Aldose Sugar Dehydrogenase
To be Published
|
|
1QO8
 
 | | The structure of the open conformation of a flavocytochrome c3 fumarate reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C3 FUMARATE REDUCTASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bamford, V, Dobbin, P.S, Richardson, D.J, Hemmings, A.M. | | Deposit date: | 1999-11-04 | | Release date: | 2000-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Open Conformation of a Flavocytochrome C3 Fumarate Reductase.
Nat.Struct.Biol., 6, 1999
|
|
1GU6
 
 | | Structure of the Periplasmic Cytochrome c Nitrite Reductase from Escherichia coli | | Descriptor: | CALCIUM ION, CYTOCHROME C552, GLYCEROL, ... | | Authors: | Bamford, V.A, Angove, H.C, Seward, H.E, Thomson, A.J, Cole, J.A, Butt, J.N, Hemmings, A.M, Richardson, D.J. | | Deposit date: | 2002-01-24 | | Release date: | 2002-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Spectroscopy of the Periplasmic Cytochrome C Nitrite Reductase from Escherichia Coli
Biochemistry, 41, 2002
|
|
2NYA
 
 | | Crystal structure of the periplasmic nitrate reductase (NAP) from Escherichia coli | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM(VI) ION, ... | | Authors: | Jepson, B.J.N, Richardson, D.J, Hemmings, A.M. | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Spectropotentiometric and structural analysis of the periplasmic nitrate reductase from Escherichia coli
J.Biol.Chem., 282, 2007
|
|
2G8S
 
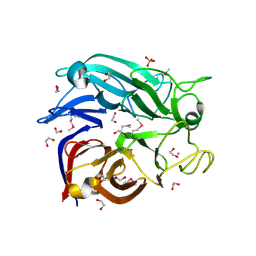 | | Crystal structure of the soluble Aldose sugar dehydrogenase (Asd) from Escherichia coli in the apo-form | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucose/sorbosone dehydrogenases, ... | | Authors: | Southall, S.M, Doel, J.J, Richardson, D.J, Oubrie, A. | | Deposit date: | 2006-03-03 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Soluble Aldose Sugar Dehydrogenase from Escherichia coli: A HIGHLY EXPOSED ACTIVE SITE CONFERRING BROAD SUBSTRATE SPECIFICITY.
J.Biol.Chem., 281, 2006
|
|
2P0B
 
 | |
2OZY
 
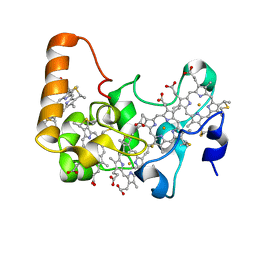 | | Crystal structure of E.coli nrfB | | Descriptor: | Cytochrome c-type protein nrfB, HEME C | | Authors: | Clarke, T.A, Richardson, D.J, Hemmings, A.M. | | Deposit date: | 2007-02-28 | | Release date: | 2007-08-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The crystal structure of the pentahaem c-type cytochrome NrfB and characterization of its solution-state interaction with the pentahaem nitrite reductase NrfA.
Biochem.J., 406, 2007
|
|
2PWZ
 
 | |
3IBE
 
 | | Crystal Structure of a Pyrazolopyrimidine Inhibitor Bound to PI3 Kinase Gamma | | Descriptor: | 1-(4-{4-morpholin-4-yl-1-[1-(pyridin-3-ylcarbonyl)piperidin-4-yl]-1H-pyrazolo[3,4-d]pyrimidin-6-yl}phenyl)-3-pyridin-4-ylurea, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Bard, J, Svenson, K. | | Deposit date: | 2009-07-15 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | ATP-Competitive Inhibitors of the Mammalian Target of Rapamycin: Design and Synthesis of Highly Potent and Selective Pyrazolopyrimidines.
J.Med.Chem., 52, 2009
|
|
4XXL
 
 | |
4LM8
 
 | |
4LMH
 
 | | Crystal structure of the outer membrane decaheme cytochrome OmcA | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, Extracellular iron oxide respiratory system surface decaheme cytochrome c component OmcA, ... | | Authors: | Edwards, M.J, Baiden, N, Clarke, T.A. | | Deposit date: | 2013-07-10 | | Release date: | 2014-04-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into electron transfer at the microbe-mineral interface: the X-ray crystal structures of Shewanella oneidensis MtrC and OmcA
FEBS Letters, 2014
|
|
4WJY
 
 | | Esherichia coli nitrite reductase NrfA H264N | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Edwards, M.J, Lockwood, C.W.J. | | Deposit date: | 2014-10-01 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Resolution of key roles for the distal pocket histidine in cytochrome C nitrite reductases.
J.Am.Chem.Soc., 137, 2015
|
|
3UCP
 
 | |
3UFH
 
 | | Crystal structure of UndA with Iron Citrate bound | | Descriptor: | CALCIUM ION, CITRATE ANION, FE (III) ION, ... | | Authors: | Edwards, M.J, Clarke, T.A. | | Deposit date: | 2011-11-01 | | Release date: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The Crystal Structure of the Extracellular 11-heme Cytochrome UndA Reveals a Conserved 10-heme Motif and Defined Binding Site for Soluble Iron Chelates.
Structure, 20, 2012
|
|
3UFK
 
 | |
3L1T
 
 | | E. coli NrfA sulfite ocmplex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Hemmings, A.M, Butt, J.N. | | Deposit date: | 2009-12-14 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and thermodynamic resolution of the interactions between sulfite and the pentahaem cytochrome NrfA from Escherichia coli
Biochem.J., 431, 2010
|
|
