2GIZ
 
 | | Structural and functional analysis of Natrin, a member of crisp-3 family blocks a variety of ion channels | | Descriptor: | Natrin-1 | | Authors: | Jiang, T, Wang, F, Li, H, Yin, C, Zhou, Y, Shu, Y, Qi, Z, Lin, Z. | | Deposit date: | 2006-03-30 | | Release date: | 2006-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and functional analysis of natrin, a venom protein that targets various ion channels
Biochem.Biophys.Res.Commun., 351, 2006
|
|
5HU9
 
 | | Crystal structure of ABL1 in complex with CHMFL-074 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-methylpiperazin-1-yl)methyl]-N-(4-methyl-3-{[1-(pyridin-3-ylcarbonyl)piperidin-4-yl]oxy}phenyl)-3-(trifluoromethyl)benzamide, CHLORIDE ION, ... | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2016-01-27 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Discovery and characterization of a novel potent type II native and mutant BCR-ABL inhibitor (CHMFL-074) for Chronic Myeloid Leukemia (CML)
Oncotarget, 7, 2016
|
|
7VO4
 
 | |
7VO5
 
 | | Pimaricin type I PKS thioesterase domain (holo Pim TE) | | Descriptor: | (1R,3S,5E,7S,11R,13E,15E,17E,19E,21R,23S,24R,25S)-11,24-dimethyl-1,3,7,21,25-pentakis(oxidanyl)-10,27-dioxabicyclo[21.3.1]heptacosa-5,13,15,17,19-pentaen-9-one, ScnS4 | | Authors: | Bai, L, Zhou, Y. | | Deposit date: | 2021-10-12 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mechanistic Insights into Chain Release of the Polyene PKS Thioesterase Domain
Acs Catalysis, 12, 2022
|
|
8TGC
 
 | |
8TG7
 
 | |
8TG8
 
 | |
8TFU
 
 | |
5GNK
 
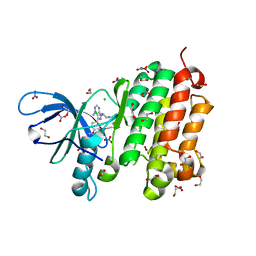 | | Crystal structure of EGFR 696-988 T790M in complex with LXX-6-34 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3R)-3-[4-azanyl-3-[3-chloranyl-4-[(1-methylimidazol-2-yl)methoxy]phenyl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]prop-2-en-1-one, CHLORIDE ION, ... | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2016-07-21 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Discovery of (R)-1-(3-(4-Amino-3-(3-chloro-4-(pyridin-2-ylmethoxy)phenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl)piperidin-1-yl)prop-2-en-1-one (CHMFL-EGFR-202) as a Novel Irreversible EGFR Mutant Kinase Inhibitor with a Distinct Binding Mode.
J. Med. Chem., 60, 2017
|
|
5GTY
 
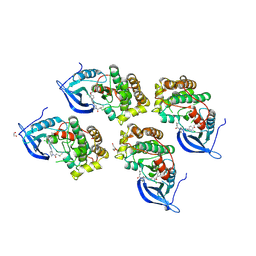 | | Crystal structure of EGFR 696-1022 T790M in complex with LXX-6-26 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3R)-3-[4-azanyl-3-[3-chloranyl-4-[(6-methylpyridin-2-yl)methoxy]phenyl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]prop-2-en-1-one, Epidermal growth factor receptor | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Discovery and characterization of a novel irreversible EGFR mutants selective and potent kinase inhibitor CHMFL-EGFR-26 with a distinct binding mode.
Oncotarget, 8, 2017
|
|
8K5N
 
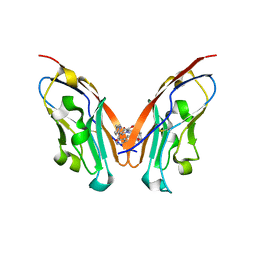 | | Discovery of Novel PD-L1 Inhibitors That Induce Dimerization and Degradation of PD-L1 Based on Fragment Coupling Strategy | | Descriptor: | 3-[(1~{S})-1-[6-methoxy-3-methyl-5-[[[(2~{S})-5-oxidanylidenepyrrolidin-2-yl]methylamino]methyl]pyridin-2-yl]oxy-2,3-dihydro-1~{H}-inden-4-yl]-2-methyl-~{N}-[5-[[[(2~{S})-5-oxidanylidenepyrrolidin-2-yl]methylamino]methyl]pyridin-2-yl]benzamide, Programmed cell death 1 ligand 1 | | Authors: | Cheng, Y, Xiao, Y.B. | | Deposit date: | 2023-07-22 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Novel PD-L1 Inhibitors That Induce the Dimerization, Internalization, and Degradation of PD-L1 Based on the Fragment Coupling Strategy.
J.Med.Chem., 66, 2023
|
|
