2VDC
 
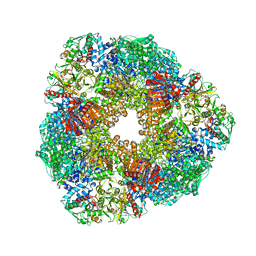 | | THE 9.5 A RESOLUTION STRUCTURE OF GLUTAMATE SYNTHASE FROM CRYO-ELECTRON MICROSCOPY AND ITS OLIGOMERIZATION BEHAVIOR IN SOLUTION: FUNCTIONAL IMPLICATIONS. | | Descriptor: | 2-OXOGLUTARIC ACID, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Cottevieille, M, Larquet, E, Jonic, S, Petoukhov, M.V, Caprini, G, Paravisi, S, Svergun, D.I, Vanoni, M.A, Boisset, N. | | Deposit date: | 2007-10-04 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | The Subnanometer Resolution Structure of the Glutamate Synthase 1.2-Mda Hexamer by Cryoelectron Microscopy and its Oligomerization Behavior in Solution: Functional Implications.
J.Biol.Chem., 283, 2008
|
|
5O5G
 
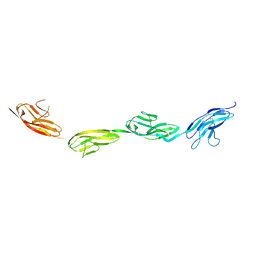 | | Robo1 Ig1 to 4 crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Roundabout homolog 1 | | Authors: | Aleksandrova, N, Gutsche, I, Kandiah, E, Avilov, S.V, Petoukhov, M.V, Seiradake, E, McCarthy, A.A. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Robo1 Forms a Compact Dimer-of-Dimers Assembly.
Structure, 26, 2018
|
|
5O5I
 
 | | Robo1 Ig5 | | Descriptor: | Roundabout homolog 1 | | Authors: | Aleksandrova, N, Gutsche, I, Kandiah, E, Avilov, S.V, Petoukhov, M.V, Seiradake, E, McCarthy, A.A. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Robo1 Forms a Compact Dimer-of-Dimers Assembly.
Structure, 26, 2018
|
|
5OPE
 
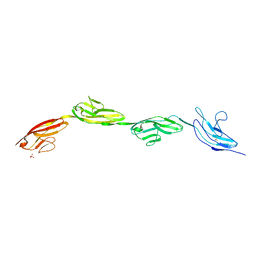 | | Robo1 Ig1-4 crystals form 2 | | Descriptor: | PHOSPHATE ION, Roundabout homolog 1 | | Authors: | Aleksandrova, N, Gutsche, I, Kandiah, E, Avilov, S.V, Petoukhov, M.V, Seiradake, E, McCarthy, A.A. | | Deposit date: | 2017-08-09 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Robo1 Forms a Compact Dimer-of-Dimers Assembly.
Structure, 26, 2018
|
|
3ZLJ
 
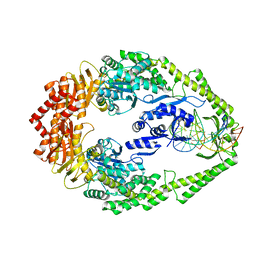 | | CRYSTAL STRUCTURE OF FULL-LENGTH E.COLI DNA MISMATCH REPAIR PROTEIN MUTS D835R MUTANT IN COMPLEX WITH GT MISMATCHED DNA | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP *AP*GP*TP*GP*TP*CP*AP)-3', 5'-D(*TP*GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*TP)-3', DNA MISMATCH REPAIR PROTEIN MUTS | | Authors: | Groothuizen, F.S, Fish, A, Petoukhov, M.V, Reumer, A, Manelyte, L, Winterwerp, H.H.K, Marinus, M.G, Lebbink, J.H.G, Svergun, D.I, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2013-02-01 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Using Stable Muts Dimers and Tetramers to Quantitatively Analyze DNA Mismatch Recognition and Sliding Clamp Formation.
Nucleic Acids Res., 41, 2013
|
|
2WVR
 
 | | Human Cdt1:Geminin complex | | Descriptor: | DNA REPLICATION FACTOR CDT1, GEMININ | | Authors: | De Marco, V, Perrakis, A. | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Quaternary Structure of the Human Cdt1-Geminin Complex Regulates DNA Replication Licensing.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5A76
 
 | |
3ECY
 
 | |
4U87
 
 | | Crystal structure of the Ba-soaked C2 crystal form of pMV158 replication initiator RepB (P3221 space group) | | Descriptor: | BARIUM ION, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Boer, D.R, Ruiz Maso, J.A, del Solar, G, Coll, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Conformational plasticity of RepB, the replication initiator protein of promiscuous streptococcal plasmid pMV158.
Sci Rep, 6, 2016
|
|
6FVE
 
 | |
6FVH
 
 | |
1OFF
 
 | |
1OFD
 
 | |
1OFE
 
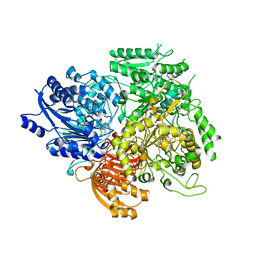 | |
4MB3
 
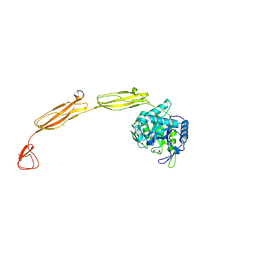 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella marina | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Chitinase 60, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3HBX
 
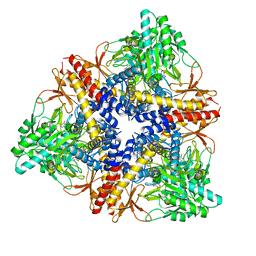 | | Crystal structure of GAD1 from Arabidopsis thaliana | | Descriptor: | Glutamate decarboxylase 1 | | Authors: | Gut, H, Dominici, P, Pilati, S, Gruetter, M.G, Capitani, G. | | Deposit date: | 2009-05-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.672 Å) | | Cite: | A common structural basis for pH- and calmodulin-mediated regulation in plant glutamate decarboxylase.
J.Mol.Biol., 392, 2009
|
|
4MB4
 
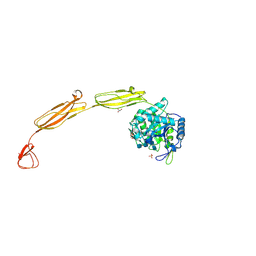 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MB5
 
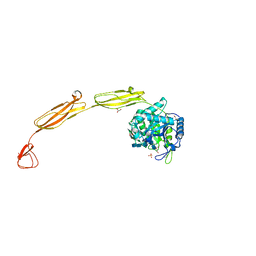 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase 60, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4ENZ
 
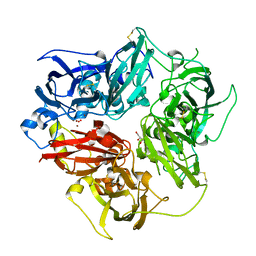 | | Structure of human ceruloplasmin at 2.6 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Samygina, V.R, Sokolov, A.V, Bourenkov, G, Vasilyev, V.B, Bartunik, H. | | Deposit date: | 2012-04-13 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ceruloplasmin: macromolecular assemblies with iron-containing acute phase proteins.
Plos One, 8, 2013
|
|
4EJX
 
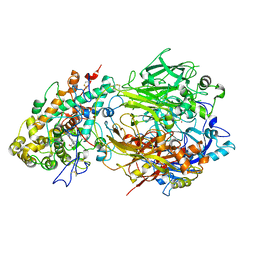 | | Structure of ceruloplasmin-myeloperoxidase complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Ceruloplasmin, ... | | Authors: | Samygina, V.R, Sokolov, A.V, Bourenkov, G, Vasilyev, V.B, Bartunik, H. | | Deposit date: | 2012-04-07 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.69 Å) | | Cite: | Ceruloplasmin: macromolecular assemblies with iron-containing acute phase proteins.
Plos One, 8, 2013
|
|
1SJR
 
 | | NMR Structure of RRM2 from Human Polypyrimidine Tract Binding Protein Isoform 1 (PTB1) | | Descriptor: | Polypyrimidine tract-binding protein 1 | | Authors: | Simpson, P.J, Monie, T.P, Szendroi, A, Davydova, N, Tyzack, J.K, Conte, M.R, Read, C.M, Cary, P.D, Svergun, D.I, Konarev, P.V, Petoukhov, M.V, Curry, S, Matthews, S.J. | | Deposit date: | 2004-03-04 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and RNA Interactions of the N-Terminal RRM Domains of PTB
Structure, 12, 2004
|
|
1SJQ
 
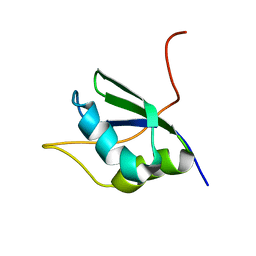 | | NMR Structure of RRM1 from Human Polypyrimidine Tract Binding Protein Isoform 1 (PTB1) | | Descriptor: | Polypyrimidine tract-binding protein 1 | | Authors: | Simpson, P.J, Monie, T.P, Szendroi, A, Davydova, N, Tyzack, J.K, Conte, M.R, Read, C.M, Cary, P.D, Svergun, D.I, Konarev, P.V, Petoukhov, M.V, Curry, S, Matthews, S.J. | | Deposit date: | 2004-03-04 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and RNA Interactions of the N-Terminal RRM Domains of PTB
Structure, 12, 2004
|
|
