2RNG
 
 | | Solution structure of big defensin | | Descriptor: | Big defensin | | Authors: | Kouno, T, Fujitani, N, Osaki, T, Kawabata, S, Nishimura, S, Mizuguchi, M, Aizawa, T, Demura, M, Nitta, K, Kawano, K. | | Deposit date: | 2007-12-27 | | Release date: | 2008-10-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A novel beta-defensin structure: a potential strategy of big defensin for overcoming resistance by Gram-positive bacteria
Biochemistry, 47, 2008
|
|
1CIX
 
 | | THREE-DIMENSIONAL STRUCTURE OF ANTIMICROBIAL PEPTIDE TACHYSTATIN A ISOLATED FROM HORSESHOE CRAB | | Descriptor: | PROTEIN (TACHYSTATIN A) | | Authors: | Fujitani, N, Kawabata, S, Osaki, T, Kumaki, Y, Demura, M, Nitta, K, Kawano, K. | | Deposit date: | 1999-04-06 | | Release date: | 2002-05-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the antimicrobial peptide tachystatin A.
J.Biol.Chem., 277, 2002
|
|
2DCW
 
 | |
2DCV
 
 | |
7F67
 
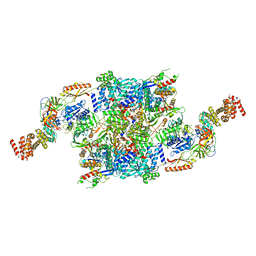 | | eIF2B-SFSV NSs-2-eIF2 | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 3, Non-structural protein NS-S, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2021-06-24 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
7F66
 
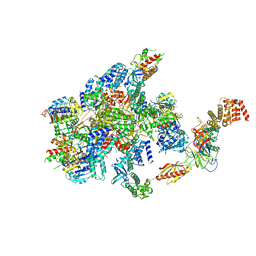 | | eIF2B-SFSV NSs-1-eIF2 | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 3, Non-structural protein NS-S, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2021-06-24 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
7F64
 
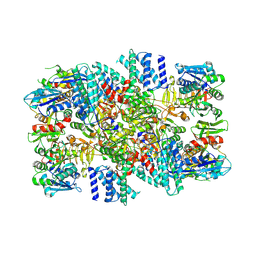 | | eIF2B-SFSV NSs | | Descriptor: | Non-structural protein NS-S, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2021-06-24 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
7VLK
 
 | | eIF2B-SFSV NSs C2-imposed | | Descriptor: | Non-structural protein NS-S, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2021-10-04 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
7E5O
 
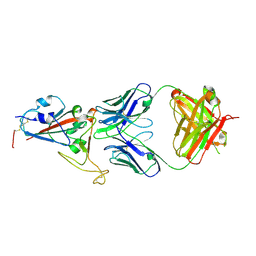 | | Crystal structure of SARS-CoV-2 RBD in complex with antibody NT-193 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NT-193 Heavy chain, NT-193 Light chain, ... | | Authors: | Kita, S, Onodera, T, Adachi, Y, Moriayma, S, Nomura, T, Tadokoro, T, Anraku, Y, Yumoto, K, Tian, C, Fukuhara, H, Suzuki, T, Tonouchi, K, Sasaki, J, Sun, L, Hashiguchi, T, Takahashi, Y, Maenaka, K. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A SARS-CoV-2 antibody broadly neutralizes SARS-related coronaviruses and variants by coordinated recognition of a virus-vulnerable site.
Immunity, 54, 2021
|
|
7EA6
 
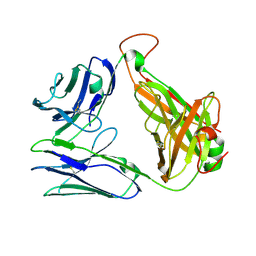 | | Crystal structure of TCR-017 ectodomain | | Descriptor: | T cell receptor 017 alpha chain, T cell receptor 017 beta chain | | Authors: | Nagae, M, Yamasaki, S. | | Deposit date: | 2021-03-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18000245 Å) | | Cite: | Identification of conserved SARS-CoV-2 spike epitopes that expand public cTfh clonotypes in mild COVID-19 patients.
J.Exp.Med., 218, 2021
|
|
7DKJ
 
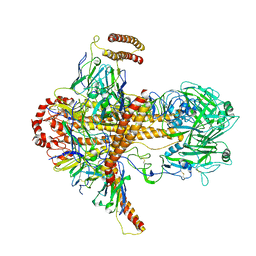 | |
2DX0
 
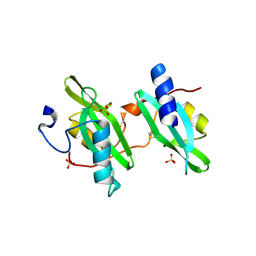 | | Crystal structure of the N-terminal SH2 domain of mouse phospholipase C-gamma 2 | | Descriptor: | Phospholipase C, gamma 2, SULFATE ION | | Authors: | Handa, N, Takagi, T, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-22 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the N-terminal SH2 domain of mouse phospholipase C-gamma 2
To be Published
|
|
