1L9H
 
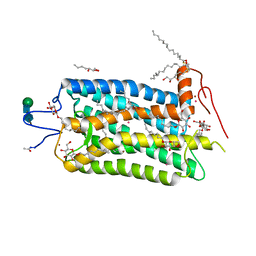 | | Crystal structure of bovine rhodopsin at 2.6 angstroms RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Okada, T, Fujiyoshi, Y, Silow, M, Navarro, J, Landau, E.M, Shichida, Y. | | Deposit date: | 2002-03-23 | | Release date: | 2002-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7YRO
 
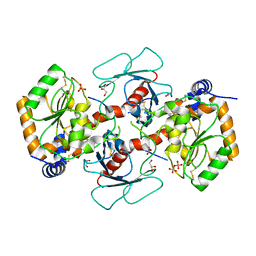 | | Crystal structure of mango fucosyltransferase 13 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, Fucosyltransferase, ... | | Authors: | Okada, T, Teramoto, T, Ihara, H, Ikeda, Y, Kakuta, Y. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of mango alpha 1,3/ alpha 1,4-fucosyltransferase elucidates unique elements that regulate Lewis A-dominant oligosaccharide assembly.
Glycobiology, 34, 2024
|
|
2E0Y
 
 | |
2E0X
 
 | |
2E0W
 
 | |
1U19
 
 | | Crystal Structure of Bovine Rhodopsin at 2.2 Angstroms Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Okada, T, Sugihara, M, Bondar, A.N, Elstner, M, Entel, P, Buss, V. | | Deposit date: | 2004-07-15 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure
J.Mol.Biol., 342, 2004
|
|
2ZYF
 
 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with magnesuim ion and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Homocitrate synthase, MAGNESIUM ION | | Authors: | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2009-01-20 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from thermus thermophilus
J.Biol.Chem., 2009
|
|
3A9I
 
 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with Lys | | Descriptor: | COBALT (II) ION, Homocitrate synthase, LYSINE | | Authors: | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2009-10-28 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from Thermus thermophilus
J.Biol.Chem., 285, 2010
|
|
2ZTK
 
 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with homocitrate | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, COPPER (II) ION, Homocitrate synthase | | Authors: | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from Thermus thermophilus
J.Biol.Chem., 285, 2010
|
|
2ZTJ
 
 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, COPPER (II) ION, Homocitrate synthase | | Authors: | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from Thermus thermophilus
J.Biol.Chem., 285, 2010
|
|
2DBW
 
 | | Crystal Structure of Gamma-glutamyltranspeptidase from Escherichia coli Acyl-Enzyme Intermediate | | Descriptor: | GAMMA-L-GLUTAMIC ACID, GLYCEROL, Gamma-glutamyltranspeptidase | | Authors: | Okada, T, Wada, K, Fukuyama, K. | | Deposit date: | 2005-12-16 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2DBX
 
 | | Crystal Structure of Gamma-glutamyltranspeptidase from Escherichia coli Complexed with L-Glutamate | | Descriptor: | CALCIUM ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Okada, T, Wada, K, Fukuyama, K. | | Deposit date: | 2005-12-16 | | Release date: | 2006-04-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DBU
 
 | |
2DG5
 
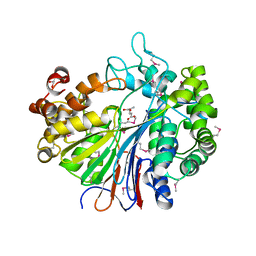 | | Crystal Structure of Gamma-glutamyl transpeptidase from Escherichia coli in complex with hydrolyzed Glutathione | | Descriptor: | CALCIUM ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Okada, T, Wada, K, Fukuyama, K. | | Deposit date: | 2006-03-08 | | Release date: | 2006-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1F88
 
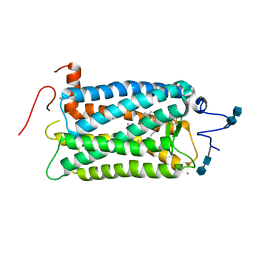 | | CRYSTAL STRUCTURE OF BOVINE RHODOPSIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MERCURY (II) ION, RETINAL, ... | | Authors: | Okada, T, Palczewski, K, Stenkamp, R.E, Miyano, M. | | Deposit date: | 2000-06-29 | | Release date: | 2000-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of rhodopsin: A G protein-coupled receptor.
Science, 289, 2000
|
|
2G87
 
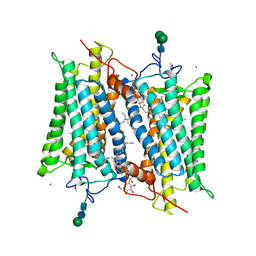 | | Crystallographic model of bathorhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Nakamichi, H, Okada, T. | | Deposit date: | 2006-03-02 | | Release date: | 2006-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic analysis of primary visual photochemistry
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
1ED7
 
 | | SOLUTION STRUCTURE OF THE CHITIN-BINDING DOMAIN OF BACILLUS CIRCULANS WL-12 CHITINASE A1 | | Descriptor: | CHITINASE A1 | | Authors: | Ikegami, T, Okada, T, Hashimoto, M, Seino, S, Watanabe, T, Shirakawa, M. | | Deposit date: | 2000-01-27 | | Release date: | 2000-05-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the chitin-binding domain of Bacillus circulans WL-12 chitinase A1.
J.Biol.Chem., 275, 2000
|
|
3OAX
 
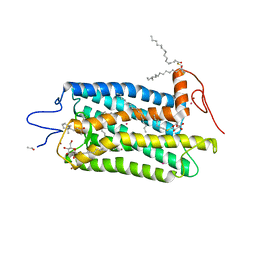 | | Crystal structure of bovine rhodopsin with beta-ionone | | Descriptor: | (3E)-4-(2,6,6-trimethylcyclohex-1-en-1-yl)but-3-en-2-one, (4E,6E)-hexadeca-1,4,6-triene, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Makino, C.L, Riley, C.K, Looney, J, Crouch, R.K, Okada, T. | | Deposit date: | 2010-08-05 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of more than one retinoid to visual opsins
Biophys.J., 99, 2010
|
|
1BM1
 
 | | CRYSTAL STRUCTURE OF BACTERIORHODOPSIN IN THE LIGHT-ADAPTED STATE | | Descriptor: | BACTERIORHODOPSIN, PHOSPHORIC ACID 2,3-BIS-(3,7,11,15-TETRAMETHYL-HEXADECYLOXY)-PROPYL ESTER 2-HYDROXO-3-PHOSPHONOOXY-PROPYL ESTER, RETINAL | | Authors: | Sato, H, Takeda, K, Tani, K, Hino, T, Okada, T, Nakasako, M, Kamiya, N, Kouyama, T. | | Deposit date: | 1998-07-28 | | Release date: | 1999-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Specific lipid-protein interactions in a novel honeycomb lattice structure of bacteriorhodopsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2PED
 
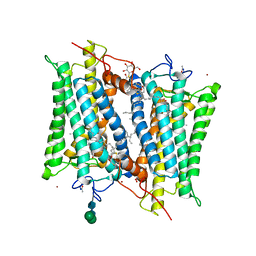 | | Crystallographic model of 9-cis-rhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Nakamichi, H, Okada, T. | | Deposit date: | 2007-04-02 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Photoisomerization mechanism of rhodopsin and 9-cis-rhodopsin revealed by x-ray crystallography
Biophys.J., 92, 2007
|
|
2HPY
 
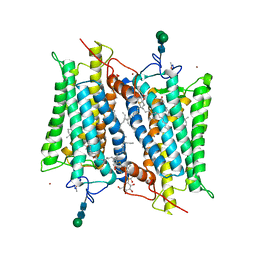 | | Crystallographic model of lumirhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Nakamichi, H, Okada, T. | | Deposit date: | 2006-07-18 | | Release date: | 2006-08-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Local peptide movement in the photoreaction intermediate of rhodopsin
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2NQO
 
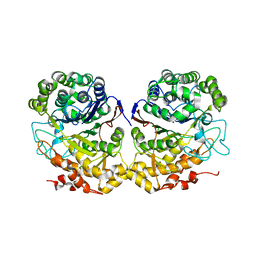 | | Crystal Structure of Helicobacter pylori gamma-Glutamyltranspeptidase | | Descriptor: | Gamma-glutamyltranspeptidase | | Authors: | Boanca, G, Sand, A, Okada, T, Suzuki, H, Kumagai, H, Fukuyama, K, Barycki, J.J. | | Deposit date: | 2006-10-31 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Autoprocessing of Helicobacter pylori gamma-glutamyltranspeptidase leads to the formation of a threonine-threonine catalytic dyad.
J.Biol.Chem., 282, 2007
|
|
3UON
 
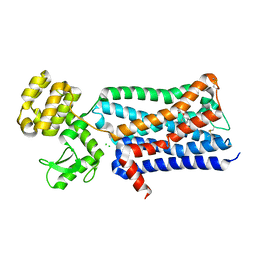 | | Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist | | Descriptor: | (3R)-1-azabicyclo[2.2.2]oct-3-yl hydroxy(diphenyl)acetate, CHLORIDE ION, Human M2 muscarinic acetylcholine, ... | | Authors: | Haga, K, Kruse, A.C, Asada, H, Yurugi-Kobayashi, T, Shiroishi, M, Zhang, C, Weis, W.I, Okada, T, Kobilka, B.K, Haga, T, Kobayashi, T. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist.
Nature, 482, 2012
|
|
6LT5
 
 | | Lysozyme protected by alginate gel | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Tomoike, F, Morita, S, Nagae, T, Okada, T. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Post-crystallization protection of protein crystals
To Be Published
|
|
7VCG
 
 | |
