1V2X
 
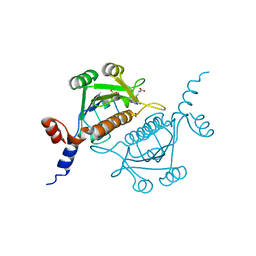 | | TrmH | | Descriptor: | PHOSPHATE ION, S-ADENOSYLMETHIONINE, tRNA (Gm18) methyltransferase | | Authors: | Nureki, O, Watanabe, K, Fukai, S, Ishii, R, Endo, Y, Hori, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-17 | | Release date: | 2004-05-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Deep Knot Structure for Construction of Active Site and Cofactor Binding Site of tRNA Modification Enzyme
STRUCTURE, 12, 2004
|
|
1GLN
 
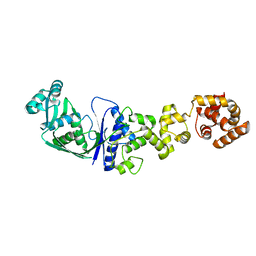 | | ARCHITECTURES OF CLASS-DEFINING AND SPECIFIC DOMAINS OF GLUTAMYL-TRNA SYNTHETASE | | Descriptor: | GLUTAMYL-TRNA SYNTHETASE | | Authors: | Nureki, O, Vassylyev, D.G, Katayanagi, K, Shimizu, T, Sekine, S, Kigawa, T, Miyazawa, T, Yokoyama, S, Morikawa, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1994-07-20 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architectures of class-defining and specific domains of glutamyl-tRNA synthetase.
Science, 267, 1995
|
|
1IPA
 
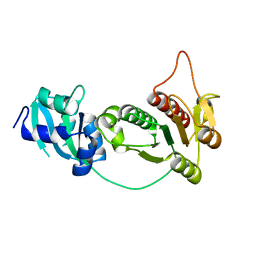 | | CRYSTAL STRUCTURE OF RNA 2'-O RIBOSE METHYLTRANSFERASE | | Descriptor: | RNA 2'-O-RIBOSE METHYLTRANSFERASE | | Authors: | Nureki, O, Shirouzu, M, Hashimoto, K, Ishitani, R, Terada, T, Tamakoshi, M, Oshima, T, Chijimatsu, M, Takio, K, Vassylyev, D.G, Shibata, T, Inoue, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-02 | | Release date: | 2002-07-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An enzyme with a deep trefoil knot for the active-site architecture.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1ILE
 
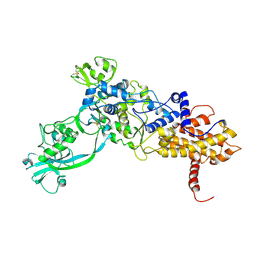 | | ISOLEUCYL-TRNA SYNTHETASE | | Descriptor: | ISOLEUCYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Nureki, O, Vassylyev, D.G, Tateno, M, Shimada, A, Nakama, T, Fukai, S, Konno, M, Schimmel, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-02-24 | | Release date: | 1999-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzyme structure with two catalytic sites for double-sieve selection of substrate.
Science, 280, 1998
|
|
3AII
 
 | |
1UEV
 
 | |
1UET
 
 | |
1UEU
 
 | |
2D6F
 
 | |
1IRX
 
 | | Crystal structure of class I lysyl-tRNA synthetase | | Descriptor: | ZINC ION, lysyl-tRNA synthetase | | Authors: | Nureki, O, Terada, T, Ishitani, R, Ambrogelly, A, Ibba, M, Soll, D, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-10-25 | | Release date: | 2002-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional convergence of two lysyl-tRNA synthetases with unrelated topologies.
Nat.Struct.Biol., 9, 2002
|
|
2ZY9
 
 | |
1N77
 
 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with tRNA(Glu) and ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glutamyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-13 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
1N78
 
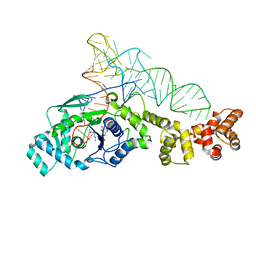 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with tRNA(Glu) and glutamol-AMP. | | Descriptor: | GLUTAMOL-AMP, Glutamyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-13 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
1N75
 
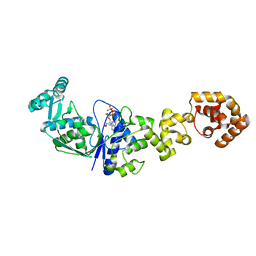 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glutamyl-tRNA synthetase, MAGNESIUM ION | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-12 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
4WIB
 
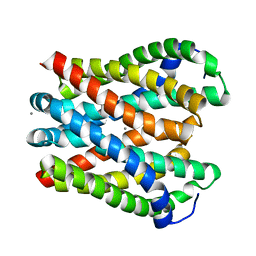 | | Crystal structure of Magnesium transporter MgtE | | Descriptor: | CALCIUM ION, Magnesium transporter MgtE | | Authors: | Takeda, H, Hattori, M, Nishizawa, T, Yamashita, K, Shah, S.T.A, Caffrey, M, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-09-25 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for ion selectivity revealed by high-resolution crystal structure of Mg(2+) channel MgtE
Nat Commun, 5, 2014
|
|
1KN0
 
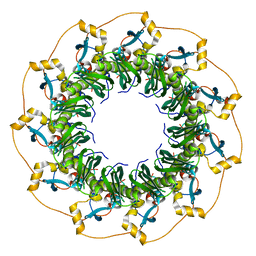 | | Crystal Structure of the human Rad52 protein | | Descriptor: | Rad52 | | Authors: | Kagawa, W, Kurumizaka, H, Ishitani, R, Fukai, S, Nureki, O, Shibata, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-12-18 | | Release date: | 2002-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form.
Mol.Cell, 10, 2002
|
|
3S7T
 
 | |
8HIN
 
 | | Structure of human SGLT2-MAP17 complex with Phlorizin | | Descriptor: | 1-[2-[(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,6-bis(oxidanyl)phenyl]-3-(4-hydroxyphenyl)propan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, PDZK1-interacting protein 1, ... | | Authors: | Hiraizumi, M, Kishida, H, Miyaguchi, I, Nureki, O. | | Deposit date: | 2022-11-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Transport and inhibition mechanism of the human SGLT2-MAP17 glucose transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
9IZR
 
 | | Cryo-EM structure of CasLambda2-crRNA-target DNA ternary complex in the NTS-cleaving state | | Descriptor: | 2-DEOXY-ADENOSINE -5'-THIO-MONOPHOSPHATE, 2-DEOXY-CYTIDINE-5'-THIOPHOSPHORATE, Cas Lambda2, ... | | Authors: | Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-08-01 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for target DNA cleavage and guide RNA processing by CRISPR-Cas lambda 2.
Commun Biol, 8, 2025
|
|
9IZS
 
 | | Cryo-EM structure of CasLambda2-crRNA-target DNA ternary complex in the TS-cleaving state | | Descriptor: | Cas Lambda2, DNA (40-MER), MAGNESIUM ION, ... | | Authors: | Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-08-01 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis for target DNA cleavage and guide RNA processing by CRISPR-Cas lambda 2.
Commun Biol, 8, 2025
|
|
9J7W
 
 | | Channel Rhodospin from Klebsormidium nitens (KnChR) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, KnChR, RETINAL | | Authors: | Wang, Y.Z, Akasaka, H, Tanaka, T, Sano, F.K, Shihoya, W, Nureki, O. | | Deposit date: | 2024-08-20 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Cryo-EM structure of a blue-shifted channelrhodopsin from Klebsormidium nitens.
Nat Commun, 16, 2025
|
|
9IZP
 
 | | Cryo-EM structure of CasLambda2-crRNA-target DNA ternary complex in the incompetent state | | Descriptor: | Cas Lambda2, DNA (40-MER), MAGNESIUM ION, ... | | Authors: | Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-08-01 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural basis for target DNA cleavage and guide RNA processing by CRISPR-Cas lambda 2.
Commun Biol, 8, 2025
|
|
9IZM
 
 | | Cryo-EM structure of CasLambda2-crRNA binary complex | | Descriptor: | Cas Lambda2, MAGNESIUM ION, RNA (58-MER) | | Authors: | Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-08-01 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis for target DNA cleavage and guide RNA processing by CRISPR-Cas lambda 2.
Commun Biol, 8, 2025
|
|
9IZQ
 
 | | Cryo-EM structure of CasLambda2-crRNA-target DNA ternary complex in the intermediate state | | Descriptor: | Cas Lambda2, DNA (40-MER), MAGNESIUM ION, ... | | Authors: | Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-08-01 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for target DNA cleavage and guide RNA processing by CRISPR-Cas lambda 2.
Commun Biol, 8, 2025
|
|
5JYJ
 
 | | Crystal structure of mouse JUNO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sperm-egg fusion protein Juno | | Authors: | Kato, K, Nishimasu, H, Morita, J, Ishitani, R, Nureki, O. | | Deposit date: | 2016-05-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the egg IZUMO1 receptor JUNO
To Be Published
|
|
