1RMS
 
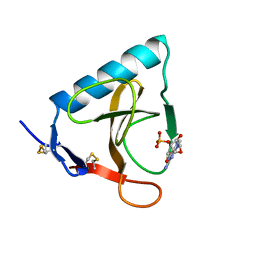 | | CRYSTAL STRUCTURES OF RIBONUCLEASE MS COMPLEXED WITH 3'-GUANYLIC ACID A GP*C ANALOGUE, 2'-DEOXY-2'-FLUOROGUANYLYL-3',5'-CYTIDINE | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE MS | | Authors: | Nonaka, T, Mitsui, Y, Nakamura, K.T. | | Deposit date: | 1991-12-02 | | Release date: | 1992-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ribonuclease Ms (as a ribonuclease T1 homologue) complexed with a guanylyl-3',5'-cytidine analogue.
Biochemistry, 32, 1993
|
|
1RDS
 
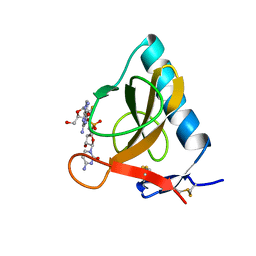 | | CRYSTAL STRUCTURE OF RIBONUCLEASE MS (AS RIBONUCLEASE T1 HOMOLOGUE) COMPLEXED WITH A GUANYLYL-3',5'-CYTIDINE ANALOGUE | | Descriptor: | 2'-FLUOROGUANYLYL-(3'-5')-PHOSPHOCYTIDINE, RIBONUCLEASE MS | | Authors: | Nonaka, T, Nakamura, K.T, Mitsui, Y. | | Deposit date: | 1993-05-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of ribonuclease Ms (as a ribonuclease T1 homologue) complexed with a guanylyl-3',5'-cytidine analogue.
Biochemistry, 32, 1993
|
|
2CVO
 
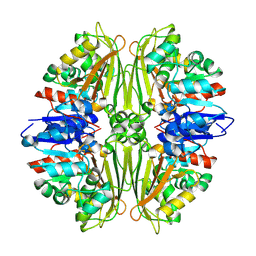 | | Crystal structure of putative N-acetyl-gamma-glutamyl-phosphate reductase (AK071544) from rice (Oryza sativa) | | Descriptor: | putative Semialdehyde dehydrogenase | | Authors: | Nonaka, T, Kita, A, Miura-Ohnuma, J, Katoh, E, Inagaki, N, Yamazaki, T, Miki, K. | | Deposit date: | 2005-06-10 | | Release date: | 2005-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative N-acetyl-gamma-glutamyl-phosphate reductase (AK071544) from rice (Oryza sativa)
Proteins, 61, 2005
|
|
1UD3
 
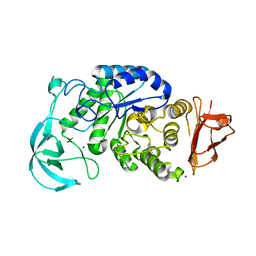 | | Crystal structure of AmyK38 N289H mutant | | Descriptor: | SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD8
 
 | | Crystal structure of AmyK38 with lithium ion | | Descriptor: | SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD4
 
 | | Crystal structure of calcium free alpha amylase from Bacillus sp. strain KSM-K38 (AmyK38, in calcium containing solution) | | Descriptor: | SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD2
 
 | | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) | | Descriptor: | GLYCEROL, SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UD5
 
 | | Crystal structure of AmyK38 with rubidium ion | | Descriptor: | RUBIDIUM ION, SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1WMF
 
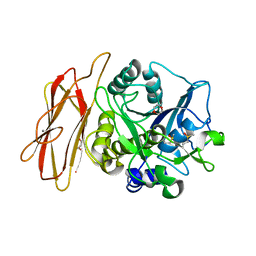 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (oxidized form, 1.73 angstrom) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CALCIUM ION, GLYCEROL, ... | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | Deposit date: | 2004-07-08 | | Release date: | 2004-09-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
1WME
 
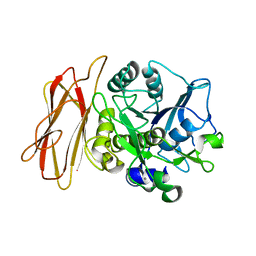 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (1.50 angstrom, 293 K) | | Descriptor: | CALCIUM ION, protease | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | Deposit date: | 2004-07-08 | | Release date: | 2004-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
1WMD
 
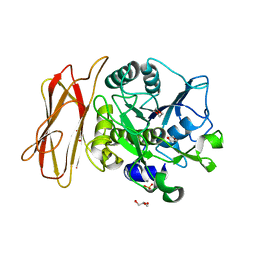 | | Crystal Structure of alkaline serine protease KP-43 from Bacillus sp. KSM-KP43 (1.30 angstrom, 100 K) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CALCIUM ION, GLYCEROL, ... | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Saeki, K, Ito, S, Horikoshi, K, Miki, K. | | Deposit date: | 2004-07-08 | | Release date: | 2004-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Crystal Structure of an Oxidatively Stable Subtilisin-like Alkaline Serine Protease, KP-43, with a C-terminal {beta}-Barrel Domain
J.Biol.Chem., 279, 2004
|
|
1UD6
 
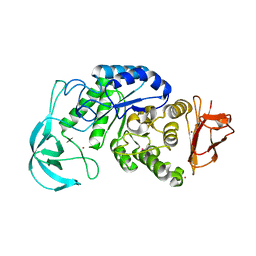 | | Crystal structure of AmyK38 with potassium ion | | Descriptor: | POTASSIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
6HFB
 
 | | Outward-facing conformation of a multidrug resistance MATE family transporter of the MOP superfamily. | | Descriptor: | CESIUM ION, Uncharacterized protein | | Authors: | Nonaka, T, Zakrzewska, S, Safarian, S, Michel, H. | | Deposit date: | 2018-08-21 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Inward-facing conformation of a multidrug resistance MATE family transporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3IWR
 
 | | Crystal structure of class I chitinase from Oryza sativa L. japonica | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chitinase | | Authors: | Kezuka, Y, Watanabe, T, Nonaka, T. | | Deposit date: | 2009-09-03 | | Release date: | 2010-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of full-length class I chitinase from rice revealed by X-ray crystallography and small-angle X-ray scattering.
Proteins, 78, 2010
|
|
5XEO
 
 | |
5XEN
 
 | |
5XEM
 
 | |
1ITX
 
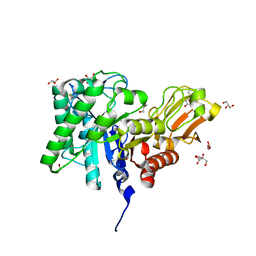 | | Catalytic Domain of Chitinase A1 from Bacillus circulans WL-12 | | Descriptor: | GLYCEROL, Glycosyl Hydrolase | | Authors: | Iwahori, F, Matsumoto, T, Watanabe, T, Nonaka, T. | | Deposit date: | 2002-02-13 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Three-dimensional structure of the catalytic domain of chitinase A1 from Bacillus circulans WL-12 at a very high resolution
PROC.JPN.ACAD.,SER.B, 75, 1999
|
|
1AHH
 
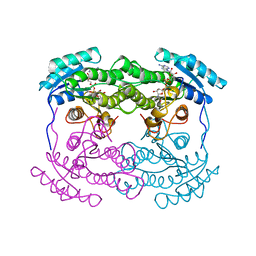 | | 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE COMPLEXED WITH NAD+ | | Descriptor: | 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tanaka, N, Nonaka, T, Mitsui, Y. | | Deposit date: | 1995-08-25 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the binary and ternary complexes of 7 alpha-hydroxysteroid dehydrogenase from Escherichia coli.
Biochemistry, 35, 1996
|
|
1AHI
 
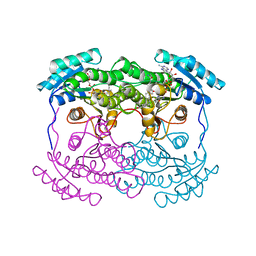 | | 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE COMPLEXED WITH NADH AND 7-OXO GLYCOCHENODEOXYCHOLIC ACID | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE, GLYCOCHENODEOXYCHOLIC ACID | | Authors: | Tanaka, N, Nonaka, T, Mitsui, Y. | | Deposit date: | 1995-08-25 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the binary and ternary complexes of 7 alpha-hydroxysteroid dehydrogenase from Escherichia coli.
Biochemistry, 35, 1996
|
|
1KWC
 
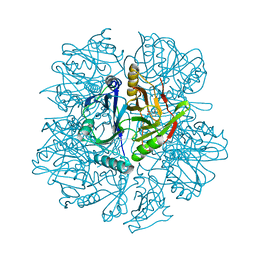 | | The His145Ala mutant of 2,3-dihydroxybiphenyl dioxygenase in complex with 2,3-dihydroxybiphenyl | | Descriptor: | 2,3-dihydroxybiphenyl dioxygenase, BIPHENYL-2,3-DIOL | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-29 | | Release date: | 2003-01-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
1KW9
 
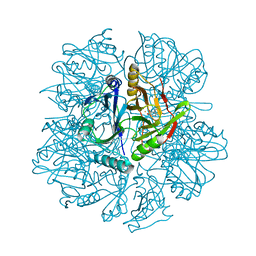 | | Crystal structure of 2,3-dihydroxybiphenyl dioxygenase (BphC) in complex with 2,3-dihydroxybiphenyl at 2.0A resolution | | Descriptor: | 2,3-Dihydroxybiphenyl dioxygenase, BIPHENYL-2,3-DIOL, FE (II) ION | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-29 | | Release date: | 2003-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
1KWB
 
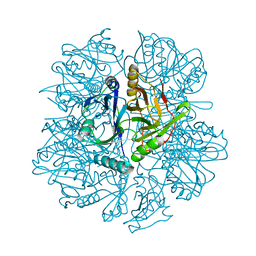 | | Crystal structure of the His145Ala mutant of 2,3-dihydroxybipheny dioxygenase (BphC) | | Descriptor: | 2,3-Dihydroxybiphenyl dioxygenase | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-29 | | Release date: | 2003-01-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
1KW8
 
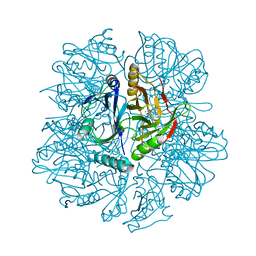 | | Crystal structure of BphC-2,3-dihydroxybiphenyl-NO complex | | Descriptor: | 2,3-Dihydroxybiphenyl dioxygenase, BIPHENYL-2,3-DIOL, FE (II) ION, ... | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-29 | | Release date: | 2003-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
4MLB
 
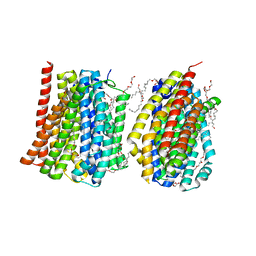 | | Reverse polarity of binding pocket suggests different function of a MOP superfamily transporter from Pyrococcus furiosus Vc1 (DSM3638) | | Descriptor: | CHLORIDE ION, PENTAETHYLENE GLYCOL MONODECYL ETHER, PF0708 | | Authors: | Malviya, V.N, Nonaka, T, Muenke, C, Koepke, J, Michel, H. | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Inward-facing conformation of a multidrug resistance MATE family transporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
