1VBH
 
 | | Pyruvate Phosphate Dikinase with bound Mg-PEP from Maize | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE, SULFATE ION, ... | | Authors: | Nakanishi, T, Nakatsu, T, Matsuoka, M, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of pyruvate phosphate dikinase from maize revealed an alternative conformation in the swiveling-domain motion
Biochemistry, 44, 2005
|
|
1VBG
 
 | | Pyruvate Phosphate Dikinase from Maize | | Descriptor: | MAGNESIUM ION, SULFATE ION, pyruvate,orthophosphate dikinase | | Authors: | Nakanishi, T, Nakatsu, T, Matsuoka, M, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of pyruvate phosphate dikinase from maize revealed an alternative conformation in the swiveling-domain motion
Biochemistry, 44, 2005
|
|
5AYU
 
 | | Crystal structure of HyHEL-10 Fv | | Descriptor: | GLYCEROL, Ig VH,anti-lysozyme, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Nakanishi, T, Tsumoto, K, Yokota, A, Kondo, H, Kumagai, I. | | Deposit date: | 2015-09-04 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of HyHEL-10 Fv
To Be Published
|
|
1WFZ
 
 | | Solution structure of Iron-sulfur cluster protein U (IscU) | | Descriptor: | ZINC ION, nitrogen fixation cluster-like | | Authors: | Nakanishi, T, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2004-11-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Iron-sulfur cluster protein U (IscU)
To be Published
|
|
1V61
 
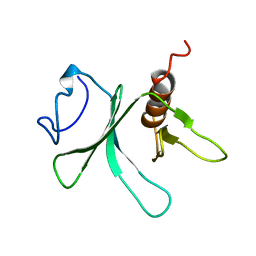 | | Solution Structure of the Pleckstrin Homology Domain of alpha-Pix | | Descriptor: | Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 | | Authors: | Nakanishi, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-26 | | Release date: | 2004-05-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Pleckstrin Homology Domain of alpha-Pix
To be Published
|
|
1WGE
 
 | | Solution structure of the mouse DESR1 | | Descriptor: | Hypothetical protein 2610018L09Rik, ZINC ION | | Authors: | Nakanishi, T, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the mouse DESR1
To be Published
|
|
1WFY
 
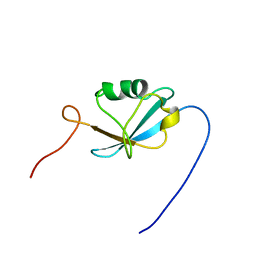 | | Solution structure of the Ras-binding domain of mouse RGS14 | | Descriptor: | regulator of G-protein signaling 14; rap1/rap2 interacting protein | | Authors: | Nakanishi, T, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2004-11-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras-binding domain of mouse RGS14
To be Published
|
|
1WGD
 
 | | Solution structure of the Ubl-domain of Herp | | Descriptor: | Homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member 1 protein | | Authors: | Nakanishi, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ubl-domain of Herp
To be Published
|
|
1WH0
 
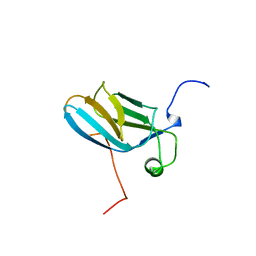 | | Solution structure of the CS domain of human USP19 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 19 | | Authors: | Nakanishi, T, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CS domain of human USP19
To be Published
|
|
1WHD
 
 | | Solution structure of the PDZ domain of RGS3 | | Descriptor: | regulator of G-protein signaling 3 | | Authors: | Nakanishi, T, Nemoto, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PDZ domain of RGS3
To be Published
|
|
2EIZ
 
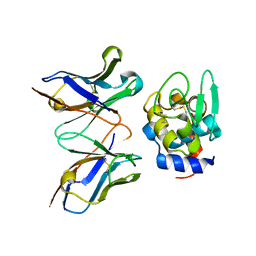 | | Crystal structure of humanized HYHEL-10 fv mutant(HW47Y)-hen lysozyme complex | | Descriptor: | ANTI-LYSOZYME ANTIBODY FV REGION, Lysozyme C | | Authors: | Nakanishi, T, Tsumoto, K, Yokota, A, Kondo, H, Kumagai, I. | | Deposit date: | 2007-03-14 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Critical contribution of VH-VL interaction to reshaping of an antibody: the case of humanization of anti-lysozyme antibody, HyHEL-10
Protein Sci., 17, 2008
|
|
2EKS
 
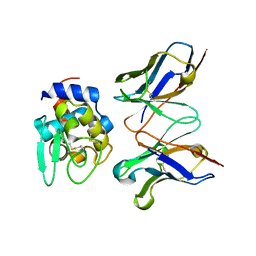 | | Crystal structure of humanized HyHEL-10 FV-HEN lysozyme complex | | Descriptor: | ANTI-LYSOZYME ANTIBODY FV REGION, Lysozyme C | | Authors: | Nakanishi, T, Tsumoto, K, Yokota, A, Kondo, H, Kumagai, I. | | Deposit date: | 2007-03-24 | | Release date: | 2008-03-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Critical contribution of VH-VL interaction to reshaping of an antibody: the case of humanization of anti-lysozyme antibody, HyHEL-10
Protein Sci., 17, 2008
|
|
2YSS
 
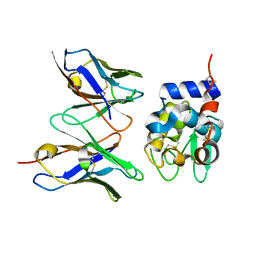 | | Crystal structure of Humanized HYHEL-10 FV mutant(HQ39KW47Y)-HEN lysozyme complex | | Descriptor: | ANTI-LYSOZYME ANTIBODY FV REGION, Lysozyme C | | Authors: | Nakanishi, T, Tsumoto, K, Yokota, A, Kondo, H, Kumagai, I. | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Critical contribution of VH-VL interaction to reshaping of an antibody: the case of humanization of anti-lysozyme antibody, HyHEL-10
Protein Sci., 17, 2008
|
|
3VVK
 
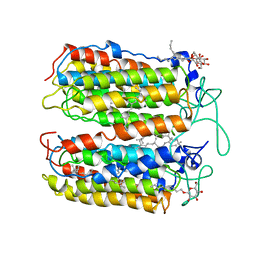 | |
3A6R
 
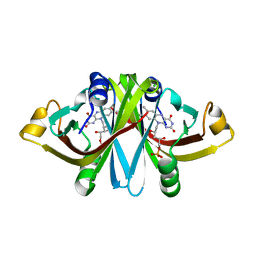 | | E13Q mutant of FMN-binding protein from Desulfovibrio vulgaris (Miyazaki F) | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, FMN-binding protein | | Authors: | Nakanishi, T, Haruyama, Y, Inoue, H, Kitamura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Effects of the disappearance of one charge on ultrafast fluorescence dynamics of the FMN binding protein.
J.Phys.Chem.B, 114, 2010
|
|
3A6Q
 
 | | E13T mutant of FMN-binding protein from Desulfovibrio vulgaris (Miyazaki F) | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, FMN-binding protein | | Authors: | Nakanishi, T, Haruyama, Y, Inoue, H, Kitamura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Effects of the disappearance of one charge on ultrafast fluorescence dynamics of the FMN binding protein.
J.Phys.Chem.B, 114, 2010
|
|
2Z4Q
 
 | | Crystal structure of a murine antibody FAB 528 | | Descriptor: | CADMIUM ION, CHLORIDE ION, anti egfr antibody fab, ... | | Authors: | Nakanishi, T, Tsumoto, K, Asano, R, Kondo, H, Kumagai, I. | | Deposit date: | 2007-06-22 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thermodynamic consequences of mutations in vernier zone residues of a humanized anti-human epidermal growth factor receptor murine antibody, 528
J.Biol.Chem., 283, 2008
|
|
5XED
 
 | | Heterodimer constructed from M61A PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
2RQH
 
 | | Structure of GSPT1/ERF3A-PABC | | Descriptor: | G1 to S phase transition 1, Polyadenylate-binding protein 1 | | Authors: | Osawa, M, Nakanishi, T, Hosoda, N, Uchida, S, Hoshino, T, Katada, I, Shimada, I. | | Deposit date: | 2009-05-08 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic Translation Termination Factor Gspt/Erf3 Recognizes Pabp with Chemical Exchange Using Two Overlapping Motifs
To be Published
|
|
2RQG
 
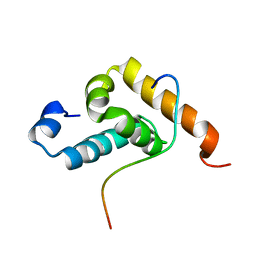 | | Structure of GSPT1/ERF3A-PABC | | Descriptor: | G1 to S phase transition 1, Polyadenylate-binding protein 1 | | Authors: | Osawa, M, Nakanishi, T, Hosoda, N, Uchida, S, Hoshino, T, Katada, I, Shimada, I. | | Deposit date: | 2009-05-08 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic Translation Termination Factor Gspt/Erf3 Recognizes Pabp with Chemical Exchange Using Two Overlapping Motifs
To be Published
|
|
7EG5
 
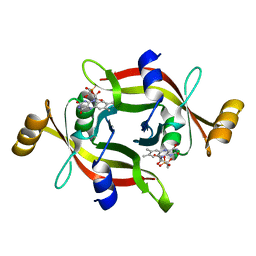 | |
5XEC
 
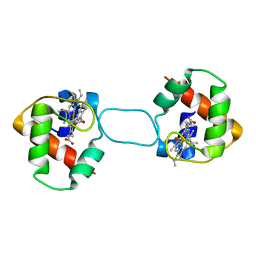 | | Heterodimer constructed from PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
5B3K
 
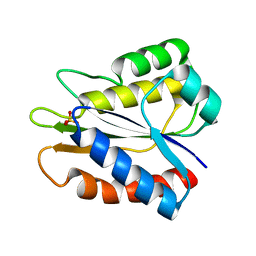 | |
5B3L
 
 | |
3VY2
 
 | |
