1BJ3
 
 | | CRYSTAL STRUCTURE OF COAGULATION FACTOR IX-BINDING PROTEIN (IX-BP) FROM VENOM OF HABU SNAKE WITH A HETERODIMER OF C-TYPE LECTIN DOMAINS | | Descriptor: | CALCIUM ION, PROTEIN (COAGULATION FACTOR IX-BINDING PROTEIN A), PROTEIN (COAGULATION FACTOR IX-BINDING PROTEIN B) | | Authors: | Mizuno, H, Fujimoto, Z, Koizumi, M, Kano, H, Atoda, H, Morita, T. | | Deposit date: | 1998-07-02 | | Release date: | 1999-08-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of coagulation factor IX-binding protein from habu snake venom at 2.6 A: implication of central loop swapping based on deletion in the linker region.
J.Mol.Biol., 289, 1999
|
|
1IXX
 
 | | CRYSTAL STRUCTURE OF COAGULATION FACTORS IX/X-BINDING PROTEIN (IX/X-BP) FROM VENOM OF HABU SNAKE WITH A HETERODIMER OF C-TYPE LECTIN DOMAINS | | Descriptor: | CALCIUM ION, COAGULATION FACTORS IX/X-BINDING PROTEIN | | Authors: | Mizuno, H, Fujimoto, Z, Koizumi, M, Kano, H. | | Deposit date: | 1997-05-01 | | Release date: | 1998-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of coagulation factors IX/X-binding protein, a heterodimer of C-type lectin domains.
Nat.Struct.Biol., 4, 1997
|
|
1IOD
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN THE COAGULATION FACTOR X BINDING PROTEIN FROM SNAKE VENOM AND THE GLA DOMAIN OF FACTOR X | | Descriptor: | CALCIUM ION, COAGULATION FACTOR X BINDING PROTEIN, COAGULATION FACTOR X GLA DOMAIN | | Authors: | Mizuno, H, Fujimoto, Z, Atoda, H, Morita, T. | | Deposit date: | 2001-02-27 | | Release date: | 2001-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an anticoagulant protein in complex with the Gla domain of factor X.
Proc.Natl.Acad.Sci.Usa, 98, 2001
|
|
2Z6Z
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-08-09 | | Release date: | 2008-07-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Z6X
 
 | | Crystal structure of 22G, the wild-type protein of the photoswitchable GFP-like protein Dronpa | | Descriptor: | photochromic protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-08-09 | | Release date: | 2008-07-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Z6Y
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-08-09 | | Release date: | 2008-07-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Z1O
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-05-10 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1PE6
 
 | | REFINED X-RAY STRUCTURE OF PAPAIN(DOT)E-64-C COMPLEX AT 2.1-ANGSTROMS RESOLUTION | | Descriptor: | METHANOL, N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-2-METHYL-BUTANE, PAPAIN | | Authors: | Yamamoto, D, Matsumoto, K, Ohishi, H, Ishida, T, Inoue, M, Kitamura, K, Mizuno, H. | | Deposit date: | 1991-05-14 | | Release date: | 1993-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined x-ray structure of papain.E-64-c complex at 2.1-A resolution.
J.Biol.Chem., 266, 1991
|
|
1UA7
 
 | | Crystal Structure Analysis of Alpha-Amylase from Bacillus Subtilis complexed with Acarbose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha-amylase, ... | | Authors: | Kagawa, M, Fujimoto, Z, Momma, M, Takase, K, Mizuno, H. | | Deposit date: | 2003-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of Bacillus subtilis alpha-amylase in complex with acarbose
J.BACTERIOL., 185, 2003
|
|
1BAG
 
 | | ALPHA-AMYLASE FROM BACILLUS SUBTILIS COMPLEXED WITH MALTOPENTAOSE | | Descriptor: | ALPHA-1,4-GLUCAN-4-GLUCANOHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Fujimoto, Z, Mizuno, H, Takase, K, Doui, N. | | Deposit date: | 1998-01-30 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a catalytic-site mutant alpha-amylase from Bacillus subtilis complexed with maltopentaose.
J.Mol.Biol., 277, 1998
|
|
2NLI
 
 | | Crystal Structure of the complex between L-lactate oxidase and a substrate analogue at 1.59 angstrom resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, HYDROGEN PEROXIDE, LACTIC ACID, ... | | Authors: | Furuichi, M, Suzuki, N, Balasundaresan, D, Yoshida, Y, Minagawa, H, Watanabe, Y, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | Deposit date: | 2006-10-20 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
1UAS
 
 | | Crystal structure of rice alpha-galactosidase | | Descriptor: | GLYCEROL, PLATINUM (II) ION, SULFATE ION, ... | | Authors: | Fujimoto, Z, Kaneko, S, Momma, M, Kobayashi, H, Mizuno, H. | | Deposit date: | 2003-03-18 | | Release date: | 2003-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of rice alpha-galactosidase complexed with D-galactose
J.Biol.Chem., 278, 2003
|
|
1UF2
 
 | | The Atomic Structure of Rice dwarf Virus (RDV) | | Descriptor: | Core protein P3, Outer capsid protein P8, Structural protein P7 | | Authors: | Nakagawa, A, Miyazaki, N, Taka, J, Naitow, H, Ogawa, A, Fujimoto, Z, Mizuno, H, Higashi, T, Watanabe, Y, Omura, T, Cheng, R.H, Tsukihara, T. | | Deposit date: | 2003-05-23 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins.
Structure, 11, 2003
|
|
4Z35
 
 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-9910539 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 3-{1-[(2S,3S)-3-(4-acetyl-3,5-dimethoxyphenyl)-2-(2,3-dihydro-1H-inden-2-ylmethyl)-3-hydroxypropyl]-4-(methoxycarbonyl)-1H-pyrrol-3-yl}propanoic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
4Z36
 
 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-3080573 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 1-(4-{[(2S,3R)-2-(2,3-dihydro-1H-inden-2-yloxy)-3-(3,5-dimethoxy-4-methylphenyl)-3-hydroxypropyl]oxy}phenyl)cyclopropan ecarboxylic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
1ISW
 
 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylobiose | | Descriptor: | beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, endo-1,4-beta-D-xylanase | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
4Z34
 
 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO9780307 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, Lysophosphatidic acid receptor 1, Soluble cytochrome b562, ... | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
1J35
 
 | | Crystal Structure of Ca(II)-bound Gla Domain of Factor IX Complexed with Binding Protein | | Descriptor: | CALCIUM ION, Coagulation factor IX, Coagulation factor IX-binding protein B chain, ... | | Authors: | Shikamoto, Y, Morita, T, Fujimoto, Z, Mizuno, H. | | Deposit date: | 2003-01-20 | | Release date: | 2003-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Mg2+- and Ca2+-bound Gla Domain of Factor IX Complexed with Binding Protein
J.Biol.Chem., 278, 2003
|
|
1ISZ
 
 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with galactose | | Descriptor: | beta-D-galactopyranose, endo-1,4-beta-D-xylanase | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
1ISX
 
 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylotriose | | Descriptor: | beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
1IT0
 
 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, endo-1,4-beta-D-xylanase | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
1ISV
 
 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylose | | Descriptor: | beta-D-xylopyranose, endo-1,4-beta-D-xylanase | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
1J34
 
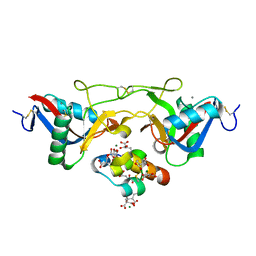 | | Crystal Structure of Mg(II)-and Ca(II)-bound Gla Domain of Factor IX Complexed with Binding Protein | | Descriptor: | CALCIUM ION, Coagulation factor IX, MAGNESIUM ION, ... | | Authors: | Shikamoto, Y, Morita, T, Fujimoto, Z, Mizuno, H. | | Deposit date: | 2003-01-20 | | Release date: | 2003-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Mg2+- and Ca2+-bound Gla Domain of Factor IX Complexed with Binding Protein
J.Biol.Chem., 278, 2003
|
|
1ISY
 
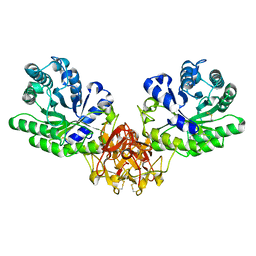 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with glucose | | Descriptor: | beta-D-glucopyranose, endo-1,4-beta-D-xylanase | | Authors: | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2001-12-27 | | Release date: | 2002-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
1C3A
 
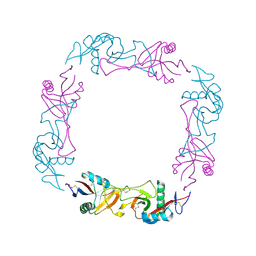 | | CRYSTAL STRUCTURE OF FLAVOCETIN-A FROM THE HABU SNAKE VENOM, A NOVEL CYCLIC TETRAMER OF C-TYPE LECTIN-LIKE HETERODIMERS | | Descriptor: | FLAVOCETIN-A: ALPHA SUBUNIT, FLAVOCETIN-A: BETA SUBUNIT | | Authors: | Fukuda, K, Mizuno, H, Atoda, H, Morita, T. | | Deposit date: | 1999-07-27 | | Release date: | 2000-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of flavocetin-A, a platelet glycoprotein Ib-binding protein, reveals a novel cyclic tetramer of C-type lectin-like heterodimers.
Biochemistry, 39, 2000
|
|
