5WX4
 
 | |
3I9T
 
 | | Crystal structure of the rat heme oxygenase (HO-1) in complex with heme binding dithiothreitol (DTT) | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Matsui, T, Unno, M, Ikeda-Saito, M. | | Deposit date: | 2009-07-13 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dioxygen activation for the self-degradation of heme: reaction mechanism and regulation of heme oxygenase.
Inorg.Chem., 49, 2010
|
|
3I9U
 
 | | Crystal structure of the rat heme oxygenase (HO-1) in complex with heme binding dithioerythritol (DTE) | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Matsui, T, Unno, M, Ikeda-Saito, M. | | Deposit date: | 2009-07-13 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dioxygen activation for the self-degradation of heme: reaction mechanism and regulation of heme oxygenase.
Inorg.Chem., 49, 2010
|
|
3I8R
 
 | | Crystal structure of the heme oxygenase from Corynebacterium diphtheriae (HmuO) in complex with heme binding ditiothreitol (DTT) | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Heme oxygenase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Matsui, T, Unno, M, Ikeda-Saito, M. | | Deposit date: | 2009-07-10 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dioxygen activation for the self-degradation of heme: reaction mechanism and regulation of heme oxygenase.
Inorg.Chem., 49, 2010
|
|
5WX6
 
 | | Alkyldiketide-CoA synthase W332Q mutant from Evodia rutaecarpa | | Descriptor: | Alkyldiketide-CoA synthase, COENZYME A, SULFATE ION | | Authors: | Matsui, T, Kodama, T, Tadakoshi, T, Morita, H. | | Deposit date: | 2017-01-06 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | 2-Alkylquinolone alkaloid biosynthesis in the medicinal plant Evodia rutaecarpa involves collaboration of two novel type III polyketide synthases
J. Biol. Chem., 292, 2017
|
|
5WX3
 
 | | Alkyldiketide-CoA synthase from Evodia rutaecarpa | | Descriptor: | Alkyldiketide-CoA synthase, COENZYME A, SULFATE ION | | Authors: | Matsui, T, Kodama, T, Tadakoshi, T, Morita, H. | | Deposit date: | 2017-01-06 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | 2-Alkylquinolone alkaloid biosynthesis in the medicinal plant Evodia rutaecarpa involves collaboration of two novel type III polyketide synthases
J. Biol. Chem., 292, 2017
|
|
5WX7
 
 | | Alkyldiketide-CoA synthase W332G mutant from Evodia rutaecarpa | | Descriptor: | Alkyldiketide-CoA synthase, COENZYME A, SULFATE ION | | Authors: | Matsui, T, Kodama, T, Tadakoshi, T, Morita, H. | | Deposit date: | 2017-01-06 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | 2-Alkylquinolone alkaloid biosynthesis in the medicinal plant Evodia rutaecarpa involves collaboration of two novel type III polyketide synthases
J. Biol. Chem., 292, 2017
|
|
5WX5
 
 | |
1WNV
 
 | | D136A mutant of Heme Oxygenase from Corynebacterium diphtheriae (HmuO) | | Descriptor: | Heme oxygenase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Matsui, T, Unno, M, Ikeda-Saito, M. | | Deposit date: | 2004-08-10 | | Release date: | 2004-11-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Roles of Distal Asp in Heme Oxygenase from Corynebacterium diphtheriae, HmuO: A WATER-DRIVEN OXYGEN ACTIVATION MECHANISM
J.Biol.Chem., 280, 2005
|
|
6AL3
 
 | | Lys49 PLA2 BPII derived from the venom of Protobothrops flavoviridis. | | Descriptor: | Basic phospholipase A2 BP-II, SULFATE ION | | Authors: | Matsui, T, Kamata, S, Suzuki, A, Oda-Ueda, N, Ogawa, T, Tanaka, Y. | | Deposit date: | 2018-09-05 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | SDS-induced oligomerization of Lys49-phospholipase A2from snake venom.
Sci Rep, 9, 2019
|
|
2DI2
 
 | | NMR structure of the HIV-2 nucleocapsid protein | | Descriptor: | Nucleocapsid protein p7, ZINC ION | | Authors: | Matsui, T, Kodera, Y, Endoh, H, Miyauchi, E, Komatsu, H, Sato, K, Tanaka, T, Kohno, T, Maeda, T. | | Deposit date: | 2006-03-27 | | Release date: | 2007-03-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | RNA Recognition Mechanism of the Minimal Active Domain of the Human Immunodeficiency Virus Type-2 Nucleocapsid Protein
J.Biochem.(Tokyo), 141, 2007
|
|
2E1X
 
 | | NMR structure of the HIV-2 nucleocapsid protein | | Descriptor: | Gag-Pol polyprotein (Pr160Gag-Pol), ZINC ION | | Authors: | Matsui, T, Kodera, Y, Miyauchi, E, Tanaka, H, Endoh, H, Komatsu, H, Tanaka, T, Kohno, T, Maeda, T. | | Deposit date: | 2006-11-03 | | Release date: | 2007-06-05 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural role of the secondary active domain of HIV-2 NCp8 in multi-functionality
Biochem.Biophys.Res.Commun., 358, 2007
|
|
2EC7
 
 | | Solution Structure of Human Immunodificiency Virus Type-2 Nucleocapsid Protein | | Descriptor: | Gag polyprotein (Pr55Gag), ZINC ION | | Authors: | Matsui, T, Kodera, Y, Tanaka, T, Endoh, H, Tanaka, H, Miyauchi, E, Komatsu, H, Kohno, T, Maeda, T. | | Deposit date: | 2007-02-10 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The RNA recognition mechanism of human immunodeficiency virus (HIV) type 2 NCp8 is different from that of HIV-1 NCp7
Biochemistry, 48, 2009
|
|
3VO8
 
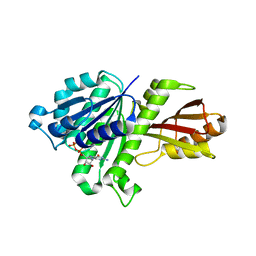 | | Staphylococcus aureus FtsZ GDP-form | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Matsui, T, Mogi, N, Yao, M, Tanaka, I. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VPA
 
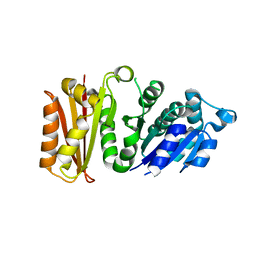 | | Staphylococcus aureus FtsZ apo-form | | Descriptor: | Cell division protein FtsZ | | Authors: | Matsui, T, Yamane, J, Mogi, N, Yao, M, Tanaka, I. | | Deposit date: | 2012-02-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VO9
 
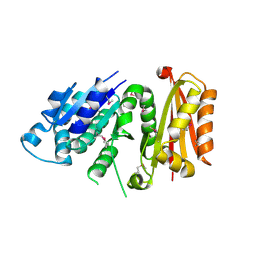 | | Staphylococcus aureus FtsZ apo-form (SeMet) | | Descriptor: | Cell division protein FtsZ | | Authors: | Matsui, T, Yamane, J, Mogi, N, Yao, M, Tanaka, I. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-29 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3WGJ
 
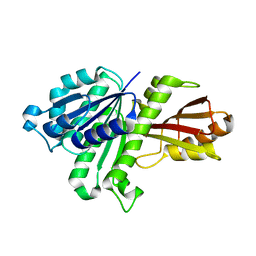 | |
3WGN
 
 | | STAPHYLOCOCCUS AUREUS FTSZ bound with GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell division protein FtsZ | | Authors: | Matsui, T, Mogi, N, Tanaka, I, Yao, M. | | Deposit date: | 2013-08-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Structural change in FtsZ Induced by intermolecular interactions between bound GTP and the T7 loop
J.Biol.Chem., 289, 2014
|
|
1VEC
 
 | | Crystal structure of the N-terminal domain of rck/p54, a human DEAD-box protein | | Descriptor: | ATP-dependent RNA helicase p54, L(+)-TARTARIC ACID, ZINC ION | | Authors: | Hogetsu, K, Matsui, T, Yukihiro, Y, Tanaka, M, Sato, T, Kumasaka, T, Tanaka, N. | | Deposit date: | 2004-03-29 | | Release date: | 2004-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insight of human DEAD-box protein rck/p54 into its substrate recognition with conformational changes
Genes Cells, 11, 2006
|
|
1WNX
 
 | | D136E mutant of Heme Oxygenase from Corynebacterium diphtheriae (HmuO) | | Descriptor: | Heme oxygenase, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Unno, M, Matsui, T, Ikeda-Saito, M. | | Deposit date: | 2004-08-10 | | Release date: | 2004-11-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Roles of Distal Asp in Heme Oxygenase from Corynebacterium diphtheriae, HmuO: A WATER-DRIVEN OXYGEN ACTIVATION MECHANISM
J.Biol.Chem., 280, 2005
|
|
1WNW
 
 | | D136N mutant of Heme Oxygenase from Corynebacterium diphtheriae (HmuO) | | Descriptor: | Heme oxygenase, IODIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Unno, M, Matsui, T, Ikeda-Saito, M. | | Deposit date: | 2004-08-10 | | Release date: | 2004-11-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Roles of Distal Asp in Heme Oxygenase from Corynebacterium diphtheriae, HmuO: A WATER-DRIVEN OXYGEN ACTIVATION MECHANISM
J.Biol.Chem., 280, 2005
|
|
3VOB
 
 | | Staphylococcus aureus FtsZ with PC190723 | | Descriptor: | 3-[(6-chloro[1,3]thiazolo[5,4-b]pyridin-2-yl)methoxy]-2,6-difluorobenzamide, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Yamane, J, Matsui, T, Mogi, N, Yamaguchi, H, Takemoto, H, Yao, M, Tanaka, I. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VOA
 
 | | Staphylococcus aureus FtsZ 12-316 GDP-form | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yamane, J, Matsui, T, Mogi, N, Yao, M, Tanaka, I. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3WVR
 
 | | Structure of ATP grasp protein with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, PGM1, ... | | Authors: | Matsui, T, Noike, M, Ooya, K, Sasaki, I, Hamano, Y, Maruyama, C, Ishikawa, J, Satoh, Y, Ito, H, Dairi, T, Morita, H. | | Deposit date: | 2014-06-04 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | A peptide ligase and the ribosome cooperate to synthesize the peptide pheganomycin.
Nat.Chem.Biol., 11, 2015
|
|
3WVQ
 
 | | Structure of ATP grasp protein | | Descriptor: | GLYCEROL, PGM1, SULFATE ION | | Authors: | Matsui, T, Noike, M, Ooya, K, Sasaki, I, Hamano, Y, Maruyama, C, Ishikawa, J, Satoh, Y, Ito, H, Dairi, T, Morita, H. | | Deposit date: | 2014-06-04 | | Release date: | 2014-11-19 | | Last modified: | 2014-12-31 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | A peptide ligase and the ribosome cooperate to synthesize the peptide pheganomycin.
Nat.Chem.Biol., 11, 2015
|
|
