2GSC
 
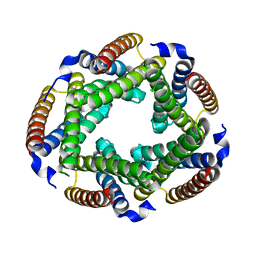 | | Crystal Structure of the Conserved Hypothetical Cytosolic Protein Xcc0516 from Xanthomonas campestris | | Descriptor: | Putative uncharacterized protein XCC0516 | | Authors: | Lin, L.Y, Ching, C.L, Chin, K.H, Chou, S.H, Chan, N.L. | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the conserved hypothetical cytosolic protein Xcc0516 from Xanthomonas campestris reveals a novel quaternary structure assembled by five four-helix bundles.
Proteins, 65, 2006
|
|
4UA2
 
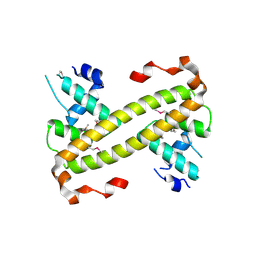 | | Crystal structure of dual function transcriptional regulator MerR from Bacillus megaterium MB1 | | Descriptor: | Regulatory protein | | Authors: | Lin, L.Y, Chang, C.C, Zou, X.W, Huang, C.C, Chan, N.L. | | Deposit date: | 2014-08-07 | | Release date: | 2015-07-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of the mercury(II)-mediated conformational switching of the dual-function transcriptional regulator MerR
Nucleic Acids Res., 43, 2015
|
|
6P45
 
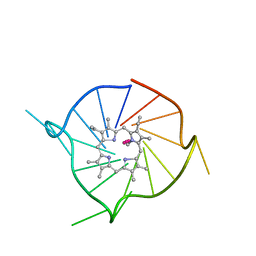 | |
6PNK
 
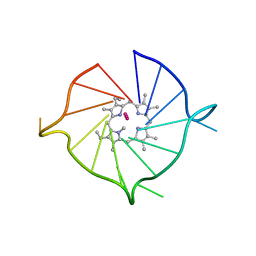 | |
3AFF
 
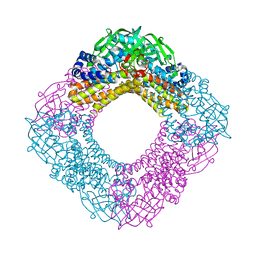 | | Crystal structure of the HsaA monooxygenase from M. tuberculosis | | Descriptor: | Hydroxylase, putative | | Authors: | D'Angelo, I, Lin, L.Y, Dresen, C, Tocheva, E.I, Strynadka, N, Eltis, L.D. | | Deposit date: | 2010-02-28 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism
J.Biol.Chem., 285, 2010
|
|
3QX3
 
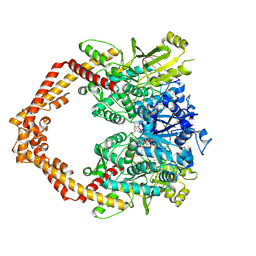 | | Human topoisomerase IIbeta in complex with DNA and etoposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*CP*TP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Wu, C.C, Li, T.K, Farh, L, Lin, L.Y, Lin, T.S, Yu, Y.J, Yen, T.J, Chiang, C.W, Chan, N.L. | | Deposit date: | 2011-03-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide
Science, 333, 2011
|
|
3AFE
 
 | | Crystal structure of the HsaA monooxygenase from M.tuberculosis | | Descriptor: | Hydroxylase, putative | | Authors: | D'Angelo, I, Lin, L.Y, Dresen, C, Tocheva, E.I, Eltis, L.D, Strynadka, N. | | Deposit date: | 2010-02-28 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism
J.Biol.Chem., 285, 2010
|
|
4UA1
 
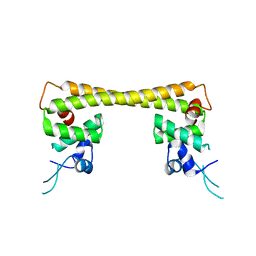 | | Crystal structure of dual function transcriptional regulator MerR form Bacillus megaterium MB1 in complex with mercury (II) ion | | Descriptor: | MERCURY (II) ION, Regulatory protein | | Authors: | Chang, C.C, Lin, L.Y, Zou, X.W, Huang, C.C, Chan, N.L. | | Deposit date: | 2014-08-07 | | Release date: | 2015-07-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis of the mercury(II)-mediated conformational switching of the dual-function transcriptional regulator MerR
Nucleic Acids Res., 43, 2015
|
|
6H2T
 
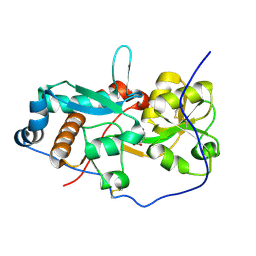 | | GlnH bound to Glu, Mycobacterium tuberculosis | | Descriptor: | GLUTAMIC ACID, Probable glutamine-binding lipoprotein GlnH (GLNBP) | | Authors: | O'Hare, H.M, Wallis, R, Lin, L.Y, Newland-Smith, Z. | | Deposit date: | 2018-07-16 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | An Aspartate-Specific Solute-Binding Protein Regulates Protein Kinase G Activity To Control Glutamate Metabolism in Mycobacteria.
MBio, 9, 2018
|
|
6H20
 
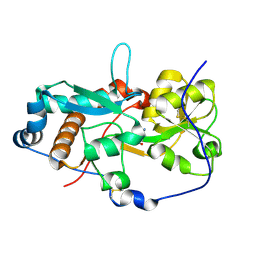 | | GlnH bound to Asn, Mycobacterium tuberculosis | | Descriptor: | ASPARAGINE, Probable glutamine-binding lipoprotein GlnH (GLNBP) | | Authors: | O'Hare, H.M, Wallis, R, Lin, L.Y, Newland-Smith, Z. | | Deposit date: | 2018-07-12 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Aspartate-Specific Solute-Binding Protein Regulates Protein Kinase G Activity To Control Glutamate Metabolism in Mycobacteria.
MBio, 9, 2018
|
|
6H1U
 
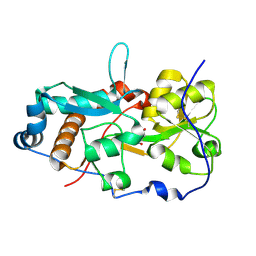 | | GlnH bound to Asp, Mycobacterium tuberculosis | | Descriptor: | ASPARTIC ACID, Probable glutamine-binding lipoprotein GlnH (GLNBP) | | Authors: | O'Hare, H.M, Wallis, R, Lin, L.Y, Newland-Smith, Z. | | Deposit date: | 2018-07-12 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | An Aspartate-Specific Solute-Binding Protein Regulates Protein Kinase G Activity To Control Glutamate Metabolism in Mycobacteria.
MBio, 9, 2018
|
|
3VMQ
 
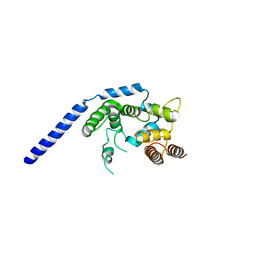 | | Crystal structure of Staphylococcus aureus membrane-bound transglycosylase: Apoenzyme | | Descriptor: | MAGNESIUM ION, Monofunctional glycosyltransferase | | Authors: | Huang, C.Y, Shih, H.W, Lin, L.Y, Tien, Y.W, Cheng, T.J.R, Cheng, W.C, Wong, C.H, Ma, C. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.518 Å) | | Cite: | Crystal structure of Staphylococcus aureus transglycosylase in complex with a lipid II analog and elucidation of peptidoglycan synthesis mechanism
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VMT
 
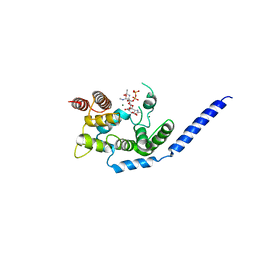 | | Crystal structure of Staphylococcus aureus membrane-bound transglycosylase in complex with a Lipid II analog | | Descriptor: | MAGNESIUM ION, Monofunctional glycosyltransferase, [(2R,3R,4R,5S,6R)-4-[(2R)-1-[[(2S)-1-[2-[2-[2-[5-[(3aS,4S,6aR)-2-oxidanylidene-1,3,3a,4,6,6a-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]ethoxy]ethoxy]ethylamino]-1-oxidanylidene-propan-2-yl]amino]-1-oxidanylidene-propan-2-yl]oxy-3-acetamido-5-[(2S,3R,4R,5R,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-(hydroxymethyl)oxan-2-yl] [oxidanyl(3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,14,18,22,26,30,34,38,42-undecaenoxy)phosphoryl] hydrogen phosphate | | Authors: | Huang, C.Y, Shih, H.W, Lin, L.Y, Tien, Y.W, Cheng, T.J.R, Cheng, W.C, Wong, C.H, Ma, C. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystal structure of Staphylococcus aureus transglycosylase in complex with a lipid II analog and elucidation of peptidoglycan synthesis mechanism
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VMR
 
 | | Crystal structure of Staphylococcus aureus membrane-bound transglycosylase in complex with moenomycin | | Descriptor: | MOENOMYCIN, Monofunctional glycosyltransferase | | Authors: | Huang, C.Y, Shih, H.W, Lin, L.Y, Tien, Y.W, Cheng, T.J.R, Cheng, W.C, Wong, C.H, Ma, C. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.688 Å) | | Cite: | Crystal structure of Staphylococcus aureus transglycosylase in complex with a lipid II analog and elucidation of peptidoglycan synthesis mechanism
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VMS
 
 | | Crystal structure of Staphylococcus aureus membrane-bound transglycosylase in complex with NBD-Lipid II | | Descriptor: | Monofunctional glycosyltransferase | | Authors: | Huang, C.Y, Shih, H.W, Lin, L.Y, Tien, Y.W, Cheng, T.J.R, Cheng, W.C, Wong, C.H, Ma, C. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Crystal structure of Staphylococcus aureus transglycosylase in complex with a lipid II analog and elucidation of peptidoglycan synthesis mechanism
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4ZGZ
 
 | |
4ZGY
 
 | |
4NPN
 
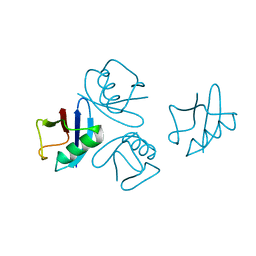 | | Crystal structure of human tetra-SUMO-2 | | Descriptor: | Small ubiquitin-related modifier 2 | | Authors: | Kung, C.C.-H, Naik, M.T, Chen, C.L, Ma, C, Huang, T.H. | | Deposit date: | 2013-11-22 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.633 Å) | | Cite: | Structural analysis of poly-SUMO chain recognition by the RNF4-SIMs domain.
Biochem.J., 462, 2014
|
|
3L7I
 
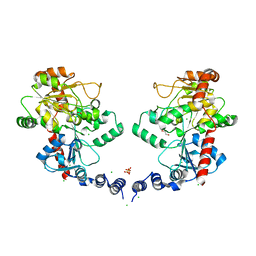 | |
3L7M
 
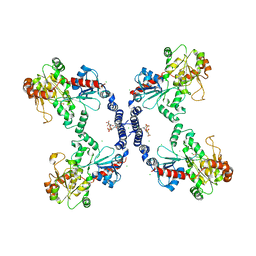 | |
6FTU
 
 | | Structure of a Quadruplex forming sequence from D. discoideum | | Descriptor: | DNA (26-MER), POTASSIUM ION | | Authors: | Guedin, A, Linda, L, Armane, S, Lacroix, L, Mergny, J.L, Thore, S, Yatsunyk, L.A. | | Deposit date: | 2018-02-23 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Quadruplexes in 'Dicty': crystal structure of a four-quartet G-quadruplex formed by G-rich motif found in the Dictyostelium discoideum genome.
Nucleic Acids Res., 46, 2018
|
|
8CXF
 
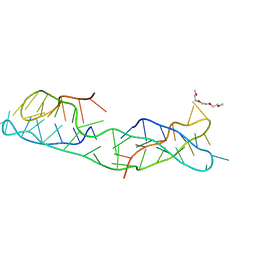 | |
8DHC
 
 | |
3L7L
 
 | | Structure of the Wall Teichoic Acid Polymerase TagF, H444N + CDPG (30 minute soak) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Lovering, A.L, Strynadka, N.C.J. | | Deposit date: | 2009-12-28 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the bacterial teichoic acid polymerase TagF provides insights into membrane association and catalysis.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L7J
 
 | |
