1FLI
 
 | | DNA-BINDING DOMAIN OF FLI-1 | | Descriptor: | FLI-1 | | Authors: | Liang, H, Mao, X, Olejniczak, E.T, Nettesheim, D.G, Yu, L, Meadows, R.P, Thompson, C.B, Fesik, S.W. | | Deposit date: | 1994-09-15 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ets domain of Fli-1 when bound to DNA.
Nat.Struct.Biol., 1, 1994
|
|
2KI0
 
 | | NMR Structure of a de novo designed beta alpha beta | | Descriptor: | DS119 | | Authors: | Liang, H, Chen, H, Fan, K, Wei, P, Guo, X, Jin, C, Zeng, C, Tang, C, Lai, L. | | Deposit date: | 2009-04-18 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | De novo design of a beta alpha beta motif.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
1DHM
 
 | | DNA-BINDING DOMAIN OF E2 FROM HUMAN PAPILLOMAVIRUS-31, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | E2 PROTEIN | | Authors: | Liang, H, Petros, A.P, Meadows, R.P, Yoon, H.S, Egan, D.A, Walter, K, Holzman, T.F, Robins, T, Fesik, S.W. | | Deposit date: | 1995-08-15 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of a human papillomavirus E2 protein: evidence for flexible DNA-binding regions.
Biochemistry, 35, 1996
|
|
8IM0
 
 | | mCherry-LaM8 complex | | Descriptor: | LaM8, MCherry fluorescent protein | | Authors: | Liang, H, Liu, R, Ding, Y. | | Deposit date: | 2023-03-05 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural Insights into the Binding of Red Fluorescent Protein mCherry-Specific Nanobodies.
Int J Mol Sci, 24, 2023
|
|
8ILX
 
 | | mCherry-LaM3 complex | | Descriptor: | LAM3, MCherry fluorescent protein | | Authors: | Liang, H, Liu, R, Ding, Y. | | Deposit date: | 2023-03-05 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural Insights into the Binding of Red Fluorescent Protein mCherry-Specific Nanobodies.
Int J Mol Sci, 24, 2023
|
|
8IM1
 
 | | mCherry-LaM1 complex | | Descriptor: | LaM1, MCherry fluorescent protein, SULFATE ION | | Authors: | Liang, H, Liu, R, Ding, Y. | | Deposit date: | 2023-03-05 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights into the Binding of Red Fluorescent Protein mCherry-Specific Nanobodies.
Int J Mol Sci, 24, 2023
|
|
7Y0Y
 
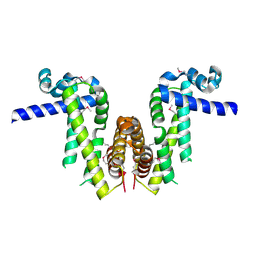 | | Crystal structure of Pseudomonas aeruginosa PvrA (SeMet) | | Descriptor: | TetR family transcriptional regulator | | Authors: | Liang, H, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
7Y0Z
 
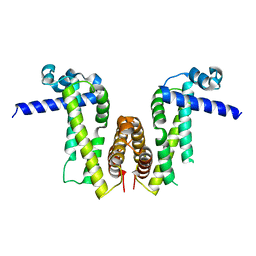 | | Crystal structure of Pseudomonas aeruginosa PvrA | | Descriptor: | TetR family transcriptional regulator | | Authors: | Liang, H, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
8HJB
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA with coenzyme A | | Descriptor: | COENZYME A, TetR family transcriptional regulator | | Authors: | Liang, H, Bartlam, M. | | Deposit date: | 2022-11-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
8YP0
 
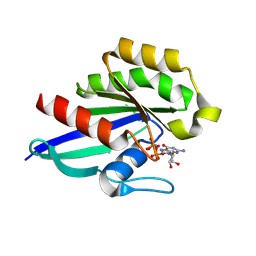 | |
8YO0
 
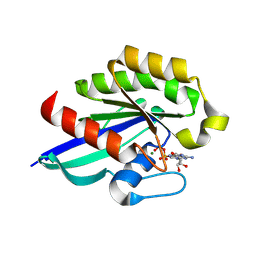 | |
1ONY
 
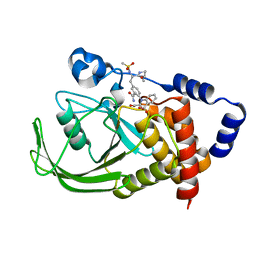 | | Oxalyl-Aryl-Amino Benzoic Acid inhibitors of PTP1B, compound 17 | | Descriptor: | 2-{[2-(2-CARBAMOYL-VINYL)-4-(2-METHANESULFONYLAMINO-2-PENTYLCARBAMOYL-ETHYL)-PHENYL]-OXALYL-AMINO}-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Liu, G, Szczepankiewicz, B.G, Pei, Z, Janowich, D.A, Xin, Z, Hadjuk, P.J, Abad-Zapatero, C, Liang, H, Hutchins, C.W, Fesik, S.W, Ballaron, S.J, Stashko, M.A, Lubben, T, Mika, A.K, Zinker, B.A, Trevillyan, J.M, Jirousek, M.R. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery and Structure-Activity Relationship of Oxalylarylaminobenzoic
Acids as Inhibitors of Protein Tyrosine Phosphatase 1B
J.Med.Chem., 46, 2003
|
|
1ONZ
 
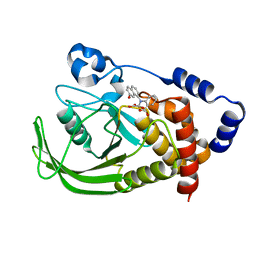 | | Oxalyl-aryl-Amino Benzoic acid Inhibitors of PTP1B, compound 8b | | Descriptor: | 2-[(7-HYDROXY-NAPHTHALEN-1-YL)-OXALYL-AMINO]-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Liu, G, Szczepankiewicz, B.G, Pei, Z, Janowich, D.A, Xin, Z, Hadjuk, P.J, Abad-Zapatero, C, Liang, H, Hutchins, C.W, Fesik, S.W, Ballaron, S.J, Stashko, M.A, Lubben, T, Mika, A.K, Zinker, B.A, Trevillyan, J.M, Jirousek, M.R. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery and Structure-Activity Relationship of Oxalylarylaminobenzoic
Acids as Inhibitors of Protein Tyrosine Phosphatase 1B
J.Med.Chem., 46, 2003
|
|
1PYN
 
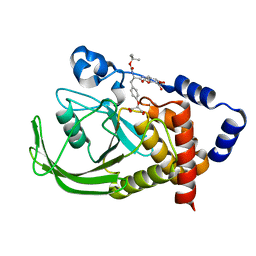 | | DUAL-SITE POTENT, SELECTIVE PROTEIN TYROSINE PHOSPHATASE 1B INHIBITOR USING A LINKED FRAGMENT STRATEGY AND A MALONATE HEAD ON THE FIRST SITE | | Descriptor: | 2-(4-{2-TERT-BUTOXYCARBONYLAMINO-2-[4-(3-HYDROXY-2-METHOXYCARBONYL-PHENOXY)-BUTYLCARBAMOYL]-ETHYL}-PHENOXY)-MALONIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Zhonghua, P, Lubben, T, Trevillyan, J.M, Stashko, M, Ballaron, S.J, Liang, H. | | Deposit date: | 2003-07-09 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and SAR of novel, potent and selective protein tyrosine phosphatase 1B inhibitors.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
4WJA
 
 | | Crystal Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Xing, M, Yang, M, Huo, W, Feng, F, Wei, L, Ning, S, Yan, Z, Li, W, Wang, Q, Hou, M, Dong, C, Guo, R, Gao, G, Ji, J, Lan, L, Liang, H, Xu, D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway.
Nat Commun, 6, 2015
|
|
5J2Y
 
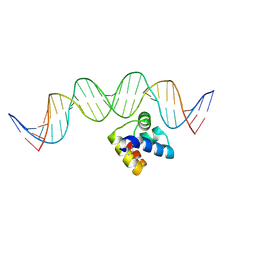 | | Molecular insight into the regulatory mechanism of the quorum-sensing repressor RsaL in Pseudomonas aeruginosa | | Descriptor: | DNA (26-MER), Regulatory protein | | Authors: | Zhao, J, Gan, J, Zhang, J, Kang, H, Kong, W, Zhu, M, Li, F, Song, Y, Qin, J, Liang, H. | | Deposit date: | 2016-03-30 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Pseudomonas aeruginosa RsaL bound to promoter DNA reaffirms its role as a global regulator involved in quorum-sensing.
Nucleic Acids Res., 45, 2017
|
|
5XF7
 
 | |
5I5O
 
 | |
5I5N
 
 | |
1NL9
 
 | | Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Compound 12 Using a Linked-Fragment Strategy | | Descriptor: | 2-{[4-(2-ACETYLAMINO-2-PENTYLCARBAMOYL-ETHYL)-NAPHTHALEN-1-YL]-OXALYL-AMINO}-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Pei, Z, Xin, Z, Lubben, T, Trevillyan, J.M, Stashko, M.A, Ballaron, S.J, Liang, H, Huang, F, Hutchins, C.W, Fesik, S.W, Jirousek, M.R. | | Deposit date: | 2003-01-06 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Selective Protein Tyrosine
Phosphatase 1B Inhibitor Using a
Linked-Fragment Strategy
J.Am.Chem.Soc., 125, 2003
|
|
1MAZ
 
 | | X-RAY STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH | | Descriptor: | Bcl-2-like protein 1 | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-09 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
1NO6
 
 | | Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Compound 5 Using a Linked-Fragment Strategy | | Descriptor: | 2-[(CARBOXYCARBONYL)(1-NAPHTHYL)AMINO]BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Pei, Z, Xin, Z, Lubben, T, Trevillyan, J.M, Stashko, M.A, Ballaron, S.J, Liang, H, Huang, F, Hutchins, C.W, Fesik, S.W, Jirousek, M.R. | | Deposit date: | 2003-01-15 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor
Using a Linked-Fragment Strategy
J.Am.Chem.Soc., 125, 2003
|
|
1NNY
 
 | | Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Compound 23 Using a Linked-Fragment Strategy | | Descriptor: | 3-({5-[(N-ACETYL-3-{4-[(CARBOXYCARBONYL)(2-CARBOXYPHENYL)AMINO]-1-NAPHTHYL}-L-ALANYL)AMINO]PENTYL}OXY)-2-NAPHTHOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Pei, Z, Xin, Z, Lubben, T, Trevillyan, J.M, Stashko, M.A, Ballaron, S.J, Liang, H, Huang, F, Hutchins, C.W, Fesik, S.W, Jirousek, M.R. | | Deposit date: | 2003-01-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor
Using a Linked-Fragment Strategy
J.Am.Chem.Soc., 125, 2003
|
|
1LXL
 
 | | NMR STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BCL-XL | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-04 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
4HQM
 
 | | The crystal structure of QsrR-menadione complex | | Descriptor: | 2-methylnaphthalene-1,4-diol, QsrR protein | | Authors: | Ji, Q, Zhang, L, Jones, M.B, Sun, F, Deng, X, Liang, H, Brugarolas, P, Gao, N, Peterson, S.N, Lan, L, Bae, T, He, C. | | Deposit date: | 2012-10-25 | | Release date: | 2013-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular mechanism of quinone signaling mediated through S-quinonization of a YodB family repressor QsrR.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
