8S1R
 
 | |
1KGD
 
 | | Crystal Structure of the Guanylate Kinase-like Domain of Human CASK | | Descriptor: | FORMIC ACID, PERIPHERAL PLASMA MEMBRANE CASK | | Authors: | Li, Y, Spangenberg, O, Paarmann, I, Konrad, M, Lavie, A. | | Deposit date: | 2001-11-26 | | Release date: | 2001-12-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.314 Å) | | Cite: | Structural basis for nucleotide-dependent regulation of membrane-associated guanylate kinase-like domains.
J.Biol.Chem., 277, 2002
|
|
9KLP
 
 | | Cryo-EM structure of ChCas12b-sgRNA-extended non-target DNA ternary complex (Complex-C) | | Descriptor: | C2c1 CRISPR-Cas endonuclease RuvC-like domain-containing protein, CALCIUM ION, Non-target DNA strand, ... | | Authors: | Li, Y, Li, J, Pei, X, Gan, J, Lin, J. | | Deposit date: | 2024-11-14 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Catalytic-state structure of Candidatus Hydrogenedentes Cas12b revealed by cryo-EM studies.
Nucleic Acids Res., 53, 2025
|
|
9KLO
 
 | | Cryo-EM structure of ChCas12b-sgRNA-extended non-target DNA ternary complex (Complex-B) | | Descriptor: | C2c1 CRISPR-Cas endonuclease RuvC-like domain-containing protein, CALCIUM ION, Non-target DNA strand, ... | | Authors: | Li, Y, Li, J, Pei, X, Gan, J, Lin, J. | | Deposit date: | 2024-11-15 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Catalytic-state structure of Candidatus Hydrogenedentes Cas12b revealed by cryo-EM studies.
Nucleic Acids Res., 53, 2025
|
|
9KLN
 
 | | Cryo-EM structure of ChCas12b-sgRNA-target DNA ternary complex (Complex-A) | | Descriptor: | C2c1 CRISPR-Cas endonuclease RuvC-like domain-containing protein, MAGNESIUM ION, Non-target DNA strand, ... | | Authors: | Li, Y, Li, J, Pei, X, Gan, J, Lin, J. | | Deposit date: | 2024-11-14 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Catalytic-state structure of Candidatus Hydrogenedentes Cas12b revealed by cryo-EM studies.
Nucleic Acids Res., 53, 2025
|
|
9KLQ
 
 | | Cryo-EM structure of ChCas12b-sgRNA-extended non-target DNA ternary complex (Complex-D) | | Descriptor: | C2c1 CRISPR-Cas endonuclease RuvC-like domain-containing protein, MAGNESIUM ION, Non-target DNA strand, ... | | Authors: | Li, Y, Li, J, Pei, X, Gan, J, Lin, J. | | Deposit date: | 2024-11-14 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Catalytic-state structure of Candidatus Hydrogenedentes Cas12b revealed by cryo-EM studies.
Nucleic Acids Res., 53, 2025
|
|
5F59
 
 | | The crystal structure of MLL3 SET domain | | Descriptor: | Histone-lysine N-methyltransferase 2C, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-04 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
5F5E
 
 | |
5F6L
 
 | | The crystal structure of MLL1 (N3861I/Q3867L) in complex with RbBP5 and Ash2L | | Descriptor: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-06 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
5F6K
 
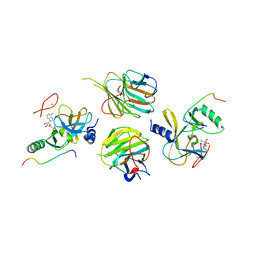 | | Crystal structure of the MLL3-Ash2L-RbBP5 complex | | Descriptor: | Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-06 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
9L5T
 
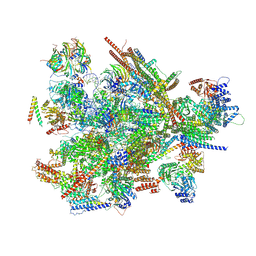 | | Cryo-EM structure of the thermophile spliceosome (state B*Q2) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Delta(14)-sterol reductase, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
9L5S
 
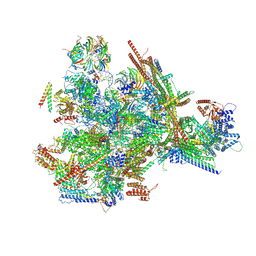 | | Cryo-EM structure of the thermophile spliceosome (state B*Q1) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Delta(14)-sterol reductase, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
9L5R
 
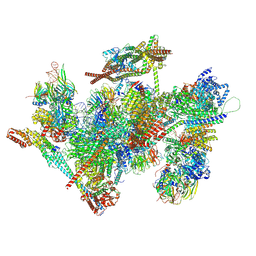 | | Cryo-EM structure of the thermophile spliceosome (state ILS) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Cell cycle control protein, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
3ETL
 
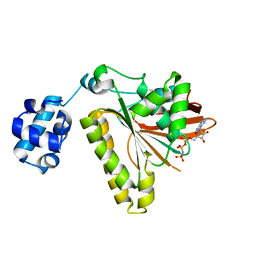 | | RadA recombinase from Methanococcus maripaludis in complex with AMPPNP | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Li, Y, He, Y, Luo, Y. | | Deposit date: | 2008-10-08 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conservation of a conformational switch in RadA recombinase from Methanococcus maripaludis.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3EWA
 
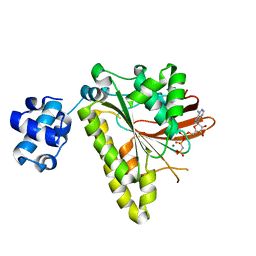 | | RADA recombinase from METHANOCOCCUS MARIPALUDIS in complex with AMPPNP and ammonium ions | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Li, Y, He, Y, Luo, Y. | | Deposit date: | 2008-10-14 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conservation of a conformational switch in RadA recombinase from Methanococcus maripaludis.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3EW9
 
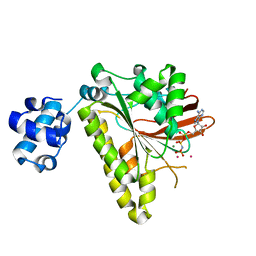 | | RADA recombinase from METHANOCOCCUS MARIPALUDIS in complex with AMPPNP and potassium ions | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, Y, He, Y, Luo, Y. | | Deposit date: | 2008-10-14 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conservation of a conformational switch in RadA recombinase from Methanococcus maripaludis.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5KTQ
 
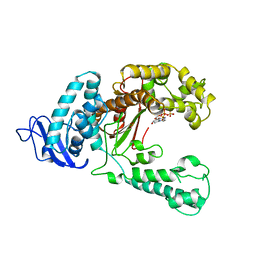 | | LARGE FRAGMENT OF TAQ DNA POLYMERASE BOUND TO DCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, PROTEIN (DNA POLYMERASE I) | | Authors: | Li, Y, Kong, Y, Korolev, S, Waksman, G. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the Klenow fragment of Thermus aquaticus DNA polymerase I complexed with deoxyribonucleoside triphosphates.
Protein Sci., 7, 1998
|
|
1HQR
 
 | | CRYSTAL STRUCTURE OF A SUPERANTIGEN BOUND TO THE HIGH-AFFINITY, ZINC-DEPENDENT SITE ON MHC CLASS II | | Descriptor: | HLA-DR ALPHA CHAIN, HLA-DR BETA CHAIN, MYELIN BASIC PROTEIN, ... | | Authors: | Li, Y, Li, H, Dimasi, N, Schlievert, P, Mariuzza, R. | | Deposit date: | 2000-12-19 | | Release date: | 2001-01-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a superantigen bound to the high-affinity, zinc-dependent site on MHC class II.
Immunity, 14, 2001
|
|
9NNR
 
 | |
9OA7
 
 | | GNAT family acetyltransferase EryM SeMet | | Descriptor: | GLYCEROL, Lysine N-acyltransferase MbtK, MAGNESIUM ION, ... | | Authors: | Li, Y, Smith, J. | | Deposit date: | 2025-04-19 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Redefining the role of the EryM acetyltransferase in natural product biosynthetic pathways.
Structure, 2025
|
|
9NNQ
 
 | | GNAT family acetyltransferase EryM | | Descriptor: | GLYCEROL, Lysine N-acyltransferase MbtK, MAGNESIUM ION, ... | | Authors: | Li, Y, Smith, J. | | Deposit date: | 2025-03-06 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Redefining the role of the EryM acetyltransferase in natural product biosynthetic pathways.
Structure, 2025
|
|
9NNS
 
 | | Flavin-dependent N5-ornithine monooxygenase EtcB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-lysine N6-monooxygenase MbtG | | Authors: | Li, Y, Liu, X, Smith, J. | | Deposit date: | 2025-03-06 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Redefining the role of the EryM acetyltransferase in natural product biosynthetic pathways.
Structure, 2025
|
|
6QNX
 
 | | Structure of the SA2/SCC1/CTCF complex | | Descriptor: | 64-kDa C-terminal product, Cohesin subunit SA-2, Transcriptional repressor CTCF | | Authors: | Li, Y, Muir, K.W, Panne, D. | | Deposit date: | 2019-02-12 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for cohesin-CTCF-anchored loops.
Nature, 578, 2020
|
|
7F68
 
 | | Crystal structure of N-ras S89D | | Descriptor: | GTPase NRas, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, Y, Sun, Q. | | Deposit date: | 2021-06-24 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of N-ras S89D
To Be Published
|
|
7SOY
 
 | | The structure of the PP2A-B56gamma1 holoenzyme-PME-1 complex | | Descriptor: | Isoform Gamma-1 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, Protein phosphatase methylesterase 1, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Li, Y, Balakrishnan, V.K, Rowse, M, Novikova, I.V, Xing, Y. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Coupling to short linear motifs creates versatile PME-1 activities in PP2A holoenzyme demethylation and inhibition.
Elife, 11, 2022
|
|
