1SK9
 
 | | Crystallographic snapshots of Aspergillus fumigatus phytase revealing its enzymatic dynamics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-phytase A, PHOSPHATE ION | | Authors: | Liu, Q, Huang, Q, Lei, X.G, Hao, Q. | | Deposit date: | 2004-03-04 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystallographic Snapshots of Aspergillus fumigatus Phytase, Revealing Its Enzymatic Dynamics
Structure, 12, 2004
|
|
1QWO
 
 | | Crystal structure of a phosphorylated phytase from Aspergillus fumigatus, revealing the structural basis for its heat resilience and catalytic pathway | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, phytase | | Authors: | Xiang, T, Liu, Q, Deacon, A.M, Koshy, M, Kriksunov, I.A, Lei, X.G, Hao, Q, Thiel, D.J. | | Deposit date: | 2003-09-03 | | Release date: | 2004-06-01 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Heat-resilient Phytase from Aspergillus fumigatus, Carrying a Phosphorylated Histidine
J.Mol.Biol., 339, 2004
|
|
1SK8
 
 | | Crystallographic snapshots of Aspergillus fumigatus phytase revealing its enzymatic dynamics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-phytase A, PHOSPHATE ION | | Authors: | Liu, Q, Huang, Q.Q, Lei, X.G, Hao, Q. | | Deposit date: | 2004-03-04 | | Release date: | 2005-02-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic Snapshots of Aspergillus fumigatus Phytase, Revealing Its Enzymatic Dynamics
Structure, 12, 2004
|
|
1SKB
 
 | | Crystallographic snapshots of Aspergillus fumigatus phytase revealing its enzymatic dynamics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-phytase A | | Authors: | Liu, Q, Huang, Q, Lei, X.G, Hao, Q. | | Deposit date: | 2004-03-04 | | Release date: | 2004-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic Snapshots of Aspergillus fumigatus Phytase, Revealing Its Enzymatic Dynamics
Structure, 12, 2004
|
|
1SKA
 
 | | Crystallographic snapshots of Aspergillus fumigatus phytase revealing its enzymatic dynamics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-phytase A | | Authors: | Liu, Q, Huang, Q, Lei, X.G, Hao, Q. | | Deposit date: | 2004-03-04 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystallographic Snapshots of Aspergillus fumigatus Phytase, Revealing Its Enzymatic Dynamics
Structure, 12, 2004
|
|
7YAV
 
 | | Crystal structure of Diels-Alderase MaDA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, MaDA1 | | Authors: | Lei, X.G, Chen, R.C, Du, X.X, Yang, J, Fan, J.P, Guo, N.X, Ding, Q. | | Deposit date: | 2022-06-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The evolutionary origin of naturally occurring intermolecular Diels-Alderases from Morus alba.
Nat Commun, 15, 2024
|
|
8ZSA
 
 | | Crystal structure of 4VPMT2 | | Descriptor: | 4-ethenylphenol, 4VPMT2, S-ADENOSYLMETHIONINE | | Authors: | Gao, L, Yang, J, Lei, X.G. | | Deposit date: | 2024-06-05 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Decoding 4-vinylanisole biosynthesis and pivotal enzymes in locusts.
Nature, 2025
|
|
7YP0
 
 | | Crystal structure of CtGST | | Descriptor: | Glutathione S-transferase | | Authors: | Yang, J, Fan, J.P, Lei, X.G. | | Deposit date: | 2022-08-02 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
7YOC
 
 | | Crystal structure of Fhb7 | | Descriptor: | Fhb7 | | Authors: | Yang, J, Liang, K, Xiao, J.Y, Lei, X.G. | | Deposit date: | 2022-08-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
8K4S
 
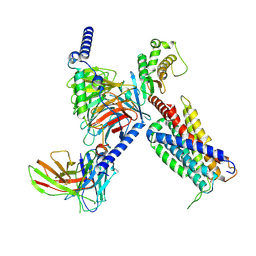 | | CryoEM structure of Gq coupled MRGPRX4 with agonist DCA-3P | | Descriptor: | (4~{R})-4-[(3~{R},5~{R},8~{R},9~{S},10~{S},12~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-12-oxidanyl-3-phosphonooxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yang, J, Fan, J.P, Lei, X.G. | | Deposit date: | 2023-07-20 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-guided discovery of bile acid derivatives for treating liver diseases without causing itch.
Cell, 187, 2024
|
|
8K2P
 
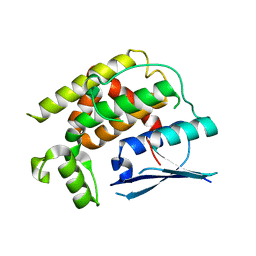 | | Crystal structure of CtGST-F76A | | Descriptor: | Glutathione S-transferase | | Authors: | Yang, J, Xiao, J.Y, Lei, X.G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
8K2O
 
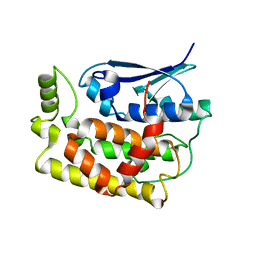 | | Crystal structure of Fhb7-M10 | | Descriptor: | Fhb7-M10 | | Authors: | Yang, J, Lei, X.G, Xiao, J.Y. | | Deposit date: | 2023-07-13 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
8KEX
 
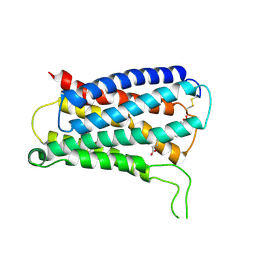 | | CryoEM structure of Gq coupled MRGPRX4 with agonist DCA-3P, local | | Descriptor: | (4~{R})-4-[(3~{R},5~{R},8~{R},9~{S},10~{S},12~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-12-oxidanyl-3-phosphonooxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Soluble cytochrome b562,Mas-related G-protein coupled receptor member X4,Green fluorescent protein | | Authors: | Yang, J, Fan, J.P, Lei, X.G. | | Deposit date: | 2023-08-14 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-guided discovery of bile acid derivatives for treating liver diseases without causing itch.
Cell, 187, 2024
|
|
6JQH
 
 | | Crystal structure of MaDA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MaDA | | Authors: | Du, X.X, Lei, X.G. | | Deposit date: | 2019-03-31 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | FAD-dependent enzyme-catalysed intermolecular [4+2] cycloaddition in natural product biosynthesis.
Nat.Chem., 12, 2020
|
|
