8QH5
 
 | | CryoEM structure of UVSSA(VHS)-CSA-DDB1-DDA1 | | Descriptor: | DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | Authors: | Lee, S.-H, Sixma, T.K. | | Deposit date: | 2023-09-06 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CryoEM structure of UVSSA(VHS)-CSA-DDB1-DDA1
To Be Published
|
|
6J2U
 
 | |
1ZLC
 
 | | Solution Conformation of alpha-conotoxin PIA | | Descriptor: | Alpha-conotoxin PIA | | Authors: | Chi, S.-W, Lee, S.-H, Kim, D.-H, Kim, J.-S, Olivera, B.M, McIntosh, J.M, Han, K.-H. | | Deposit date: | 2005-05-05 | | Release date: | 2006-05-02 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of alpha-conotoxin PIA, a novel antagonist of alpha6 subunit containing nicotinic acetylcholine receptors
Biochem.Biophys.Res.Commun., 338, 2005
|
|
5T9J
 
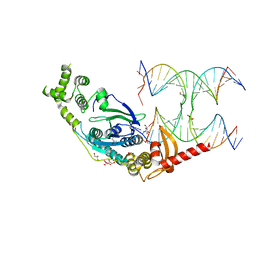 | | Crystal Structure of human GEN1 in complex with Holliday junction DNA in the upper interface | | Descriptor: | DNA (5'-D(*DAP*DCP*DGP*DAP*DTP*DGP*DGP*DAP*DGP*DCP*DCP*DGP*DCP*DTP*DAP*DGP*DGP*DCP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DAP*DTP*DTP*DCP*DCP*DGP*DGP*DAP*DTP*DTP*DAP*DGP*DGP*DGP*DAP*DTP*DGP*DC)-3'), DNA (5'-D(*DGP*DAP*DGP*DCP*DCP*DTP*DAP*DGP*DCP*DGP*DTP*DCP*DCP*DGP*DGP*DAP*DAP*DTP*DTP*DC)-3'), ... | | Authors: | Lee, S.-H, Biertumpfel, C. | | Deposit date: | 2016-09-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.00012732 Å) | | Cite: | Human Holliday junction resolvase GEN1 uses a chromodomain for efficient DNA recognition and cleavage.
Elife, 4, 2015
|
|
6ABX
 
 | |
6ABW
 
 | |
6ABV
 
 | |
6ABY
 
 | | Crystal structure of citrate synthase (Msed_1522) from Metallosphaera sedula in complex with oxaloacetate | | Descriptor: | ACETYL COENZYME *A, Citrate synthase, GLYCEROL, ... | | Authors: | Lee, S.-H, Son, H.-F, Kim, K.-J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the inhibition properties of archaeon citrate synthase from Metallosphaera sedula.
PLoS ONE, 14, 2019
|
|
8IKR
 
 | | Crystal structure of DpaA | | Descriptor: | YkuD domain-containing protein | | Authors: | Wang, H.-J, Hsieh, K.-Y, Lee, S.-H, Chang, C.-I. | | Deposit date: | 2023-03-01 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the hydrolytic activity of the transpeptidase-like protein DpaA to detach Braun's lipoprotein from peptidoglycan.
Mbio, 14, 2023
|
|
3L6Y
 
 | | Crystal structure of p120 catenin in complex with E-cadherin | | Descriptor: | Catenin delta-1, E-cadherin | | Authors: | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
3L6X
 
 | | Crystal structure of p120 catenin in complex with E-cadherin | | Descriptor: | Catenin delta-1, E-cadherin, SULFATE ION | | Authors: | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
3K9K
 
 | | Transposase domain of Metnase | | Descriptor: | Histone-lysine N-methyltransferase SETMAR | | Authors: | Goodwin, K.D, He, H, Imasaki, T, Lee, S.-H, Georgiadis, M.M. | | Deposit date: | 2009-10-15 | | Release date: | 2010-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the human Hsmar1-derived transposase domain in the DNA repair enzyme Metnase.
Biochemistry, 49, 2010
|
|
3K9J
 
 | | Transposase domain of Metnase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Histone-lysine N-methyltransferase SETMAR | | Authors: | Goodwin, K.D, He, H, Imasaki, T, Lee, S.-H, Georgiadis, M.M. | | Deposit date: | 2009-10-15 | | Release date: | 2010-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal structure of the human Hsmar1-derived transposase domain in the DNA repair enzyme Metnase.
Biochemistry, 49, 2010
|
|
1MVE
 
 | | Crystal structure of a natural circularly-permutated jellyroll protein: 1,3-1,4-beta-D-glucanase from Fibrobacter succinogenes | | Descriptor: | CALCIUM ION, Truncated 1,3-1,4-beta-D-glucanase | | Authors: | Tsai, L.-C, Shyur, L.-F, Lee, S.-H, Lin, S.-S, Yuan, H.S. | | Deposit date: | 2002-09-25 | | Release date: | 2003-07-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Natural Circularly Permuted Jellyroll Protein: 1,3-1,4-beta-D-Glucanase from Fibrobacter succinogenes.
J.Mol.Biol., 330, 2003
|
|
2G9L
 
 | |
