2D3Y
 
 | | Crystal structure of uracil-DNA glycosylase from Thermus Thermophilus HB8 | | Descriptor: | 2'-DEOXYURIDINE-5'-MONOPHOSPHATE, ACETATE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of family 5 uracil-DNA glycosylase bound to DNA.
J.Mol.Biol., 373, 2007
|
|
2DDG
 
 | | Crystal structure of uracil-DNA glycosylase in complex with AP:G containing DNA | | Descriptor: | 5'-D(*AP*TP*GP*TP*TP*GP*CP*(D1P)P*TP*TP*AP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*TP*AP*AP*GP*GP*CP*AP*AP*CP*A)-3', ACETATE ION, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Hoseki, J, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-28 | | Release date: | 2007-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of family 5 uracil-DNA glycosylase bound to DNA.
J.Mol.Biol., 373, 2007
|
|
2DP6
 
 | | Crystal structure of uracil-DNA glycosylase in complex with AP:C containing DNA | | Descriptor: | 5'-D(*AP*TP*GP*TP*TP*GP*CP*(D1P)P*TP*TP*AP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*TP*AP*AP*CP*GP*CP*AP*AP*CP*A)-3', DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Kuramitsu, S, Hoseki, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-07 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Family 5 Uracil-DNA Glycosylase Bound to DNA Reveals Insights into the Mechanism for Substrate Recognition and Catalysis
To be Published
|
|
2DEM
 
 | | Crystal structure of Uracil-DNA glycosylase in complex with AP:A containing DNA | | Descriptor: | 5'-D(*AP*TP*GP*TP*TP*GP*CP*(D1P)P*TP*TP*AP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*TP*AP*AP*AP*GP*CP*AP*AP*CP*A)-3', DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Hoseki, J, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of family 5 uracil-DNA glycosylase bound to DNA.
J.Mol.Biol., 373, 2007
|
|
5YJX
 
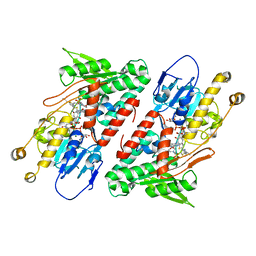 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with myxothiazol. | | Descriptor: | (2Z,6E)-7-{2'-[(2E,4E)-1,6-DIMETHYLHEPTA-2,4-DIENYL]-2,4'-BI-1,3-THIAZOL-4-YL}-3,5-DIMETHOXY-4-METHYLHEPTA-2,6-DIENAMID E, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
5YJY
 
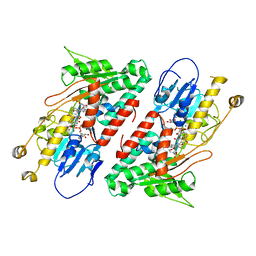 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with AC0-12. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-dodecyl-1-oxidanidyl-quinolin-1-ium-4-ol, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
5YJW
 
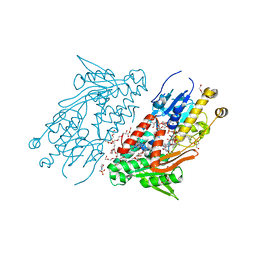 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with the competitive inhibitor, stigmatellin. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
