1YNU
 
 | | Crystal structure of apple ACC synthase in complex with L-vinylglycine | | Descriptor: | 1-aminocyclopropane-1-carboxylate synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[O-PHOSPHONOPYRIDOXYL]-AMINO- BUTYRIC ACID, ... | | Authors: | Capitani, G, Tschopp, M, Eliot, A.C, Kirsch, J.F, Grutter, M.G. | | Deposit date: | 2005-01-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of ACC synthase inactivated by the mechanism-based inhibitor L-vinylglycine.
Febs Lett., 579, 2005
|
|
3FSL
 
 | | Crystal structure of tyrosine aminotransferase tripple mutant (P181Q,R183G,A321K) from Escherichia coli at 2.35 A resolution | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Aromatic-amino-acid aminotransferase | | Authors: | Malashkevich, V.N, Ng, B, Kirsch, J.F. | | Deposit date: | 2009-01-09 | | Release date: | 2009-01-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of tyrosine aminotransferase tripple mutant (P181Q,R183G,A321K) from Escherichia coli at 2.35 A resolution
To be Published
|
|
1B8G
 
 | | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE | | Descriptor: | PROTEIN (1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Capitani, G, Hohenester, E, Feng, L, Storici, P, Kirsch, J.F, Jansonius, J.N. | | Deposit date: | 1999-01-31 | | Release date: | 2000-01-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of 1-aminocyclopropane-1-carboxylate synthase, a key enzyme in the biosynthesis of the plant hormone ethylene.
J.Mol.Biol., 294, 1999
|
|
3N4I
 
 | | Crystal structure of the SHV-1 D104E beta-lactamase/beta-lactamase inhibitor protein (BLIP) complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein | | Authors: | Hanes, M.S, Reynolds, K.A, McNamara, C, Ghosh, P, Bonomo, R.A, Kirsch, J.F, Handel, T.M. | | Deposit date: | 2010-05-21 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Specificity and Cooperativity at beta-lactamase Position 104 in TEM-1/BLIP and SHV-1/BLIP interactions
Proteins, 79, 2011
|
|
2G2W
 
 | | Crystal Structure of the SHV D104K Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein | | Authors: | Reynolds, K.A, Thomson, J.M, Corbett, K.D, Bethel, C.R, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2006-02-16 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Computational Characterization of the SHV-1 beta-Lactamase-beta-Lactamase Inhibitor Protein Interface.
J.Biol.Chem., 281, 2006
|
|
2G2U
 
 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein | | Authors: | Reynolds, K.A, Thomson, J.M, Corbett, K.D, Bethel, C.R, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2006-02-16 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Computational Characterization of the SHV-1 beta-Lactamase-beta-Lactamase Inhibitor Protein Interface.
J.Biol.Chem., 281, 2006
|
|
1TOK
 
 | | Maleic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, MALEIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOJ
 
 | | Hydrocinnamic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOI
 
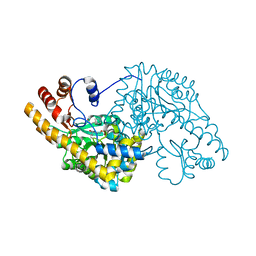 | | Hydrocinnamic acid-bound structure of Hexamutant + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOE
 
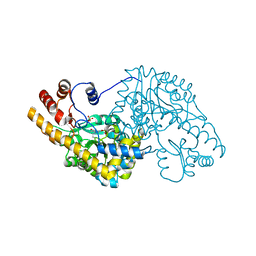 | | Unliganded structure of Hexamutant + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, SULFATE ION | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOG
 
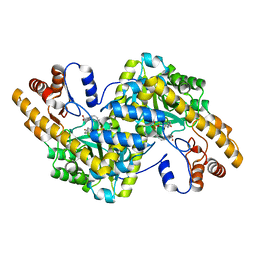 | | Hydrocinnamic acid-bound structure of SRHEPT + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1M4N
 
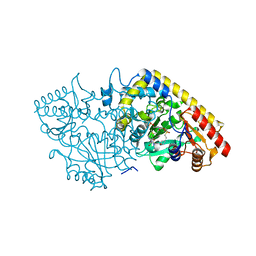 | | CRYSTAL STRUCTURE OF APPLE ACC SYNTHASE IN COMPLEX WITH [2-(AMINO-OXY)ETHYL](5'-DEOXYADENOSIN-5'-YL)(METHYL)SULFONIUM | | Descriptor: | (2-AMINOOXY-ETHYL)-[5-(6-AMINO-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDRO-FURAN-2-YLMETHYL]-METHYL-SULFONIUM, 1-aminocyclopropane-1-carboxylate synthase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Capitani, G, Eliot, A.C, Gut, H, Khomutov, R.M, Kirsch, J.F, Grutter, M.G. | | Deposit date: | 2002-07-03 | | Release date: | 2003-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of 1-aminocyclopropane-1-carboxylate synthase in complex with an amino-oxy analogue of the substrate: implications for substrate binding.
BIOCHEM.BIOPHYS.ACTA PROTEINS & PROTEOMICS, 1647, 2003
|
|
1M7Y
 
 | | Crystal structure of apple ACC synthase in complex with L-aminoethoxyvinylglycine | | Descriptor: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, 1-aminocyclopropane-1-carboxylate synthase | | Authors: | Capitani, G, McCarthy, D, Gut, H, Gruetter, M.G, Kirsch, J.F. | | Deposit date: | 2002-07-23 | | Release date: | 2002-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Apple 1-Aminocyclopropane-1-carboxylate Synthase in Complex with the Inhibitor
L-Aminoethoxyvinylglycine
J.Biol.Chem., 277, 2002
|
|
1MGV
 
 | | Crystal Structure of the R391A Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | 7,8-diamino-pelargonic acid aminotransferase, ISOPROPYL ALCOHOL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Eliot, A.C, Sandmark, J, Schneider, G, Kirsch, J.F. | | Deposit date: | 2002-08-16 | | Release date: | 2002-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Dual-Specific Active Site of 7,8-Diaminopelargonic Acid Synthase and the Effect of the R391A Mutation
Biochemistry, 41, 2002
|
|
3C4O
 
 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) E73M/S130K/S146M complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Reynolds, K.A, Hanes, M.S, Thomson, J.M, Antczak, A.J, Berger, J.M, Bonomo, R.A, Kirsch, J.F, Handel, T.M. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational redesign of the SHV-1 beta-lactamase/beta-lactamase inhibitor protein interface.
J.Mol.Biol., 382, 2008
|
|
3C4P
 
 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) E73M complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Reynolds, K.A, Hanes, M.S, Thomson, J.M, Antczak, A.J, Berger, J.M, Bonomo, R.A, Kirsch, J.F, Handel, T.M. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Computational redesign of the SHV-1 beta-lactamase/beta-lactamase inhibitor protein interface.
J.Mol.Biol., 382, 2008
|
|
1S06
 
 | | Crystal Structure of the R253K Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, SODIUM ION | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
1S08
 
 | | Crystal Structure of the D147N Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, SODIUM ION | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
1S0A
 
 | | Crystal Structure of the Y17F Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, SODIUM ION | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
1S09
 
 | | Crystal Structure of the Y144F Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, SODIUM ION | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
1S07
 
 | | Crystal Structure of the R253A Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, ISOPROPYL ALCOHOL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
1AHF
 
 | | ASPARTATE AMINOTRANSFERASE HEXAMUTANT | | Descriptor: | ASPARTATE AMINOTRANSFERASE, INDOLYLPROPIONIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Malashkevich, V.N, Jansonius, J.N. | | Deposit date: | 1995-02-22 | | Release date: | 1995-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alternating arginine-modulated substrate specificity in an engineered tyrosine aminotransferase.
Nat.Struct.Biol., 2, 1995
|
|
1AHG
 
 | | ASPARTATE AMINOTRANSFERASE HEXAMUTANT | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, TYROSINE | | Authors: | Malashkevich, V.N, Jansonius, J.N. | | Deposit date: | 1995-02-22 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alternating arginine-modulated substrate specificity in an engineered tyrosine aminotransferase.
Nat.Struct.Biol., 2, 1995
|
|
1AHY
 
 | | ASPARTATE AMINOTRANSFERASE HEXAMUTANT | | Descriptor: | ASPARTATE AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Malashkevich, V.N, Jansonius, J.N. | | Deposit date: | 1995-02-21 | | Release date: | 1995-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alternating arginine-modulated substrate specificity in an engineered tyrosine aminotransferase.
Nat.Struct.Biol., 2, 1995
|
|
1AHE
 
 | | ASPARTATE AMINOTRANSFERASE HEXAMUTANT | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Malashkevich, V.N, Jansonius, J.N. | | Deposit date: | 1995-02-22 | | Release date: | 1995-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alternating arginine-modulated substrate specificity in an engineered tyrosine aminotransferase.
Nat.Struct.Biol., 2, 1995
|
|
