4WD3
 
 | | Crystal structure of an L-amino acid ligase RizA | | Descriptor: | L-amino acid ligase | | Authors: | Kagawa, W, Arai, T, Kino, K, Kurumizaka, H. | | Deposit date: | 2014-09-06 | | Release date: | 2015-09-09 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of RizA, an L-amino-acid ligase from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4P7X
 
 | | L-pipecolic acid-bound L-proline cis-4-hydroxylase | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-OXOGLUTARIC ACID, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ... | | Authors: | Shomura, Y, Koketsu, K, Moriwaki, K, Hayashi, M, Mitsuhashi, S, Hara, R, Kino, K, Higuchi, Y. | | Deposit date: | 2014-03-28 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Refined Regio- and Stereoselective Hydroxylation of l-Pipecolic Acid by Protein Engineering of l-Proline cis-4-Hydroxylase Based on the X-ray Crystal Structure.
Acs Synth Biol, 4, 2015
|
|
4P7W
 
 | | L-proline-bound L-proline cis-4-hydroxylase | | Descriptor: | 2-OXOGLUTARIC ACID, COBALT (II) ION, L-proline cis-4-hydroxylase, ... | | Authors: | Shomura, Y, Koketsu, K, Moriwaki, K, Hayashi, M, Mitsuhashi, S, Hara, R, Kino, K, Higuchi, Y. | | Deposit date: | 2014-03-28 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined Regio- and Stereoselective Hydroxylation of l-Pipecolic Acid by Protein Engineering of l-Proline cis-4-Hydroxylase Based on the X-ray Crystal Structure.
Acs Synth Biol, 4, 2015
|
|
5ZK6
 
 | |
5ZCT
 
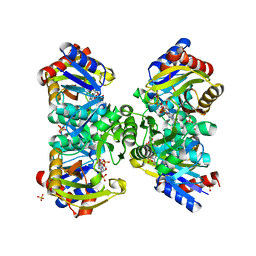 | | The crystal structure of the poly-alpha-L-glutamate peptides synthetase RimK at 2.05 angstrom resolution. | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6--L-glutamate ligase, ... | | Authors: | Arimura, Y, Kono, T, Kino, K, Kurumizaka, H. | | Deposit date: | 2018-02-20 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural polymorphism of the Escherichia coli poly-alpha-L-glutamate synthetase RimK
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3VOT
 
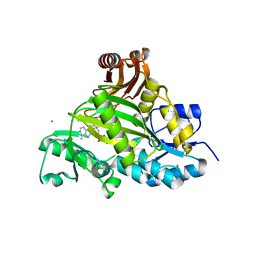 | | Crystal structure of L-amino acid ligase from Bacillus licheniformis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, M, Takahashi, Y, Noguchi, A, Arai, T, Yagasaki, M, Kino, K, Saito, J. | | Deposit date: | 2012-02-08 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of L-amino-acid ligase from Bacillus licheniformis
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1QQI
 
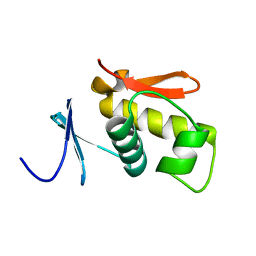 | | SOLUTION STRUCTURE OF THE DNA-BINDING AND TRANSACTIVATION DOMAIN OF PHOB FROM ESCHERICHIA COLI | | Descriptor: | PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | Authors: | Okamura, H, Hanaoka, S, Nagadoi, A, Makino, K, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-06-07 | | Release date: | 2000-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box.
J.Mol.Biol., 295, 2000
|
|
3WXQ
 
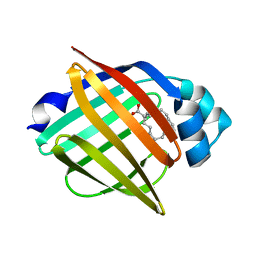 | | Serial femtosecond X-ray structure of human fatty acid-binding protein type-3 (FABP3) in complex with stearic acid (C18:0) determined using X-ray free-electron laser at SACLA | | Descriptor: | Fatty acid-binding protein, heart, STEARIC ACID | | Authors: | Mizohata, E, Suzuki, M, Kakinouchi, K, Sugiyama, S, Murata, M, Sugahara, M, Nango, E, Tanaka, T, Tanaka, R, Tono, K, Song, C, Hatsui, T, Joti, Y, Yabashi, M, Iwata, S. | | Deposit date: | 2014-08-04 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
2D42
 
 | | Crystal structure analysis of a non-toxic crystal protein from Bacillus thuringiensis | | Descriptor: | non-toxic crystal protein | | Authors: | Akiba, T, Higuchi, K, Mizuki, E, Ekino, K, Shin, T, Ohba, M, Kanai, R, Harata, K. | | Deposit date: | 2005-10-05 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Nontoxic crystal protein from Bacillus thuringiensis demonstrates a remarkable structural similarity to beta-pore-forming toxins
Proteins, 63, 2006
|
|
1UAN
 
 | | Crystal structure of the conserved protein TT1542 from Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TT1542 | | Authors: | Handa, N, Terada, T, Tame, J.R.H, Park, S.-Y, Kinoshita, K, Ota, M, Nakamura, H, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-03-12 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the conserved protein TT1542 from Thermus thermophilus HB8
PROTEIN SCI., 12, 2003
|
|
4Y9H
 
 | | The 1.43 angstrom crystal structure of bacteriorhodopsin crystallized from bicelles | | Descriptor: | Bacteriorhodopsin, DECANE, DODECANE, ... | | Authors: | Saiki, H, Sugiyama, S, Kakinouchi, K, Kawatake, S, Hanashima, S, Matsumori, N, Murata, M. | | Deposit date: | 2015-02-17 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The 1.43 angstrom crystal structure of bacteriorhodopsin crystallized from bicelles
To Be Published
|
|
5B27
 
 | | The 1.02A structure of human FABP3 M20S mutant complexed with palmitic acid | | Descriptor: | Fatty acid-binding protein, heart, PALMITIC ACID, ... | | Authors: | Matsuoka, D, Sugiyama, S, Kakinouchi, K, Niiyama, M, Murata, M, Matsuoka, S. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The 1.02A structure of human FABP3 M20S mutant complexed with palmitic acid.
To Be Published
|
|
5B29
 
 | | The 1.28A structure of human FABP3 F16V mutant complexed with palmitic acid at room temperature | | Descriptor: | Fatty acid-binding protein, heart, PALMITIC ACID | | Authors: | Matsuoka, D, Sugiyama, S, Kakinouchi, K, Niiyama, M, Murata, M, Matsuoka, S. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The 1.28A structure of human FABP3 F16V mutant complexed with palmitic acid at room temperature.
To Be Published
|
|
5B28
 
 | | The 0.90A structure of human FABP3 F16V mutant complexed with palmitic acid | | Descriptor: | Fatty acid-binding protein, heart, PALMITIC ACID, ... | | Authors: | Matsuoka, D, Sugiyama, S, Kakinouchi, K, Niiyama, M, Murata, M, Matsuoka, S. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The 0.90A structure of human FABP3 F16V mutant complexed with palmitic acid.
To Be Published
|
|
7EUW
 
 | | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing 4-[2-[1-(4-bromophenyl)-5-phenyl-1H-pyrazol-3-yl]phenoxy] (HA174) | | Descriptor: | 4-[2-[1-(4-bromophenyl)-5-phenyl-pyrazol-3-yl]phenoxy]butanoic acid, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-05-19 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray structure of the human heart fatty acid-binding protein complexed with 4-[2-[1-(4-bromophenyl)-5-phenyl-1H-pyrazol-3-yl]phenoxy] (HA176)
To Be Published
|
|
7EUV
 
 | | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing Flurbiprofen | | Descriptor: | (2R)-2-(3-fluoro-4-phenyl-phenyl)propanoic acid, Fatty acid-binding protein, heart | | Authors: | Sugiyama, S, Kakinouchi, K, Hoshina, M, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-05-19 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | X-ray structure of the human heart fatty acid-binding protein complexed with Flurbiprofen
To Be Published
|
|
7FAG
 
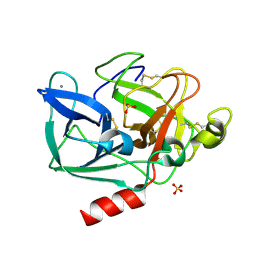 | | Room temperature structure of elastase with high-strength agarose hydrogel | | Descriptor: | CALCIUM ION, Chymotrypsin-like elastase family member 1, SULFATE ION, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Adachi, H, Murata, M, Mori, Y. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Room temperature structure of elastase with high-strength agarose hydrogel
To Be Published
|
|
7FBF
 
 | | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with octanoic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with octanoic acid
To Be Published
|
|
7FBN
 
 | | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMF solution containing dibutylhydroxytoluene (BHT) | | Descriptor: | 2,6-ditert-butyl-4-methyl-phenol, ACETIC ACID, Fatty acid-binding protein, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMF solution containing dibutylhydroxytoluene (BHT)
To Be Published
|
|
7FBM
 
 | | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing dibutylhydroxytoluene (BHT) | | Descriptor: | 2,6-ditert-butyl-4-methyl-phenol, ACETIC ACID, Fatty acid-binding protein, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing dibutylhydroxytoluene (BHT)
To Be Published
|
|
7FC4
 
 | | X-ray structure of the human heart fatty acid-binding protein complexed with S-Ibuprofen | | Descriptor: | ACETIC ACID, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-13 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing dibutylhydroxytoluene (BHT)
To Be Published
|
|
7FCG
 
 | | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing Indometacin | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing Indometacin
To Be Published
|
|
7FCX
 
 | | Room temperature structure of the human heart fatty acid-binding protein complexed with hexanoic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXANOIC ACID | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Room temperature structure of the human heart fatty acid-binding protein complexed with hexanoic acid
To Be Published
|
|
7FD7
 
 | | The 1.00 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with perfluoroheptanoic acid | | Descriptor: | Fatty acid-binding protein, heart, PENTAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Hara, T, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The 1.00 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with perfluoroheptanoic acid
To Be Published
|
|
7FDT
 
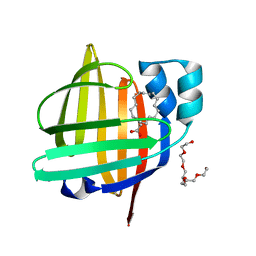 | | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with elaidic acid | | Descriptor: | 9-OCTADECENOIC ACID, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-07-17 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with octanoic acid
To Be Published
|
|
