2PNV
 
 | | Crystal Structure of the leucine zipper domain of small-conductance Ca2+-activated K+ (SKCa) channel from Rattus norvegicus | | Descriptor: | Small conductance calcium-activated potassium channel protein 2 | | Authors: | Kim, J.Y, Kim, M.K, Kang, G.B, Park, C.S, Eom, S.H. | | Deposit date: | 2007-04-25 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the leucine zipper domain of small-conductance Ca2+-activated K+ (SK(Ca)) channel from Rattus norvegicus.
Proteins, 70, 2008
|
|
9MQI
 
 | | Inactive Mu-Opioid Receptor with Nb6M, NabFab, and isoquinuclidine compound #020_E1 | | Descriptor: | Mu-type opioid receptor, NabFab Heavy Chain, NabFab Light Chain, ... | | Authors: | Kim, J.Y, Vigneron, S.F, Billesbolle, C, Manglik, A, Shoichet, B.K. | | Deposit date: | 2025-01-03 | | Release date: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Docking 14 million virtual isoquinuclidines against the mu and kappa opioid receptors reveals dual antagonists-inverse agonists with reduced withdrawal effects.
Biorxiv, 2025
|
|
9MQL
 
 | | Locally-Refined Inactive Kappa-Opioid Receptor with Nb6M, NabFab, and isoquinuclidine compound #020_E1 | | Descriptor: | Kappa-Opioid Receptor, methyl (1S,3R,4S,6S,8M)-2-[(1-ethyl-1H-pyrazol-4-yl)methyl]-8-(3-hydroxyphenyl)-3,4-dimethyl-2-azabicyclo[2.2.2]oct-7-ene-6-carboxylate | | Authors: | Kim, J.Y, Vigneron, S.F, Billesbolle, C, Manglik, A, Shoichet, B.K. | | Deposit date: | 2025-01-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Docking 14 million virtual isoquinuclidines against the mu and kappa opioid receptors reveals dual antagonists-inverse agonists with reduced withdrawal effects.
Biorxiv, 2025
|
|
9MQK
 
 | | Inactive Kappa-Opioid Receptor with Nb6M, NabFab, and isoquinuclidine compound #020_E1 | | Descriptor: | Kappa-Opioid Receptor, NabFab Heavy Chain, NabFab Light Chain, ... | | Authors: | Kim, J.Y, Vigneron, S.F, Billesbolle, C, Manglik, A, Shoichet, B.K. | | Deposit date: | 2025-01-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Docking 14 million virtual isoquinuclidines against the mu and kappa opioid receptors reveals dual antagonists-inverse agonists with reduced withdrawal effects.
Biorxiv, 2025
|
|
9MQJ
 
 | | Locally-refined Inactive Mu-Opioid Receptor with Nb6M, NabFab, and isoquinuclidine compound #020_E1 | | Descriptor: | Mu-type opioid receptor, methyl (1S,3R,4S,6S,8M)-2-[(1-ethyl-1H-pyrazol-4-yl)methyl]-8-(3-hydroxyphenyl)-3,4-dimethyl-2-azabicyclo[2.2.2]oct-7-ene-6-carboxylate | | Authors: | Kim, J.Y, Vigneron, S.F, Billesbolle, C, Manglik, A, Shoichet, B.K. | | Deposit date: | 2025-01-03 | | Release date: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Docking 14 million virtual isoquinuclidines against the mu and kappa opioid receptors reveals dual antagonists-inverse agonists with reduced withdrawal effects.
Biorxiv, 2025
|
|
9MQH
 
 | | Inactive Mu-Opioid Receptor with Nb6M, NabFab, and isoquinuclidine compound #33 | | Descriptor: | Mu-type opioid receptor | | Authors: | Kim, J.Y, Vigneron, S.F, Billesbolle, C, Manglik, A, Shoichet, B.K. | | Deposit date: | 2025-01-03 | | Release date: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Docking 14 million virtual isoquinuclidines against the mu and kappa opioid receptors reveals dual antagonists-inverse agonists with reduced withdrawal effects.
Biorxiv, 2025
|
|
1LTU
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM, APO (NO IRON BOUND) STRUCTURE | | Descriptor: | PHENYLALANINE-4-HYDROXYLASE | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
1LTZ
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM PHENYLALANINE HYDROXYLASE, STRUCTURE HAS BOUND IRON (III) AND OXIDIZED COFACTOR 7,8-DIHYDROBIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, CHLORIDE ION, FE (III) ION, ... | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-21 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
4GSU
 
 | | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Probable conserved lipoprotein LPPS | | Authors: | Kim, H.S, Kim, J, Im, H.N, Yoon, J.Y, An, D.R, Yoon, H.J, Kim, J.Y, Min, H.K, Kim, S.-J, Lee, J.Y, Han, B.W, Suh, S.W. | | Deposit date: | 2012-08-28 | | Release date: | 2013-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GSQ
 
 | | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains | | Descriptor: | CALCIUM ION, GLYCEROL, Probable conserved lipoprotein LPPS | | Authors: | Kim, H.S, Kim, J, Im, H.N, Yoon, J.Y, An, D.R, Yoon, H.J, Kim, J.Y, Min, H.K, Kim, S.-J, Lee, J.Y, Han, B.W, Suh, S.W. | | Deposit date: | 2012-08-28 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GSR
 
 | | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains | | Descriptor: | 2-(ETHYLMERCURI-THIO)-BENZOIC ACID, GLYCEROL, Probable conserved lipoprotein LPPS | | Authors: | Kim, H.S, Kim, J, Im, H.N, Yoon, J.Y, An, D.R, Yoon, H.J, Kim, J.Y, Min, H.K, Kim, S.-J, Lee, J.Y, Han, B.W, Suh, S.W. | | Deposit date: | 2012-08-28 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FYB
 
 | | Structural and functional characterizations of a thioredoxin-fold protein from Helicobacter pylori | | Descriptor: | GLYCEROL, Thiol:disulfide interchange protein (DsbC) | | Authors: | Yoon, J.Y, Kim, J, Lee, S.J, Im, H.N, Kim, H.S, Yoon, H, An, D.R, Kim, J.Y, Kim, S, Han, B.W, Suh, S.W. | | Deposit date: | 2012-07-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional characterization of HP0377, a thioredoxin-fold protein from Helicobacter pylori
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FYC
 
 | | Structural and functional characterizations of a thioredoxin-fold protein from Helicobacter pylori | | Descriptor: | TETRAETHYLENE GLYCOL, Thiol:disulfide interchange protein (DsbC) | | Authors: | Yoon, J.Y, Kim, J, Lee, S.J, Im, H.N, Kim, H.S, Yoon, H, An, D.R, Kim, J.Y, Kim, S, Han, B.W, Suh, S.W. | | Deposit date: | 2012-07-04 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural and functional characterization of HP0377, a thioredoxin-fold protein from Helicobacter pylori
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3UE8
 
 | | Kynurenine Aminotransferase II Inhibitors | | Descriptor: | (5-hydroxy-4-{[(1-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl)amino]methyl}-6-methylpyridin-3-yl)methyl dihydrogen phosphate, CHLORIDE ION, Kynurenine/alpha-aminoadipate aminotransferase, ... | | Authors: | Dounay, A.B, Anderson, M, Bechle, B.M, Campbell, B.M, Claffey, M.M, Evdokimov, A, Edelweiss, E, Fonseca, K.R, Gan, X, Ghosh, S, Hayward, M.M, Horner, W, Kim, J.Y, McAllister, L.A, Pandit, J, Paradis, V, Parikh, V.D, Reese, M.R, Rong, S.B, Salafia, M.A, Schuyten, K, Strick, C.A, Tuttle, J.B, Valentine, J, Wang, H, Zawadzke, L.E, Verhoest, P.R. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Discovery of Brain-Penetrant, Irreversible Kynurenine Aminotransferase II Inhibitors for Schizophrenia.
ACS Med Chem Lett, 3, 2012
|
|
4Q6Q
 
 | | Structural analysis of the Zn-form II of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6N
 
 | | Structural analysis of the tripeptide-bound form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | CALCIUM ION, Conserved hypothetical secreted protein, GLYCEROL, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6M
 
 | | Structural analysis of the apo-form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | CALCIUM ION, Conserved hypothetical secreted protein, GLYCEROL | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6O
 
 | | Structural analysis of the mDAP-bound form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6P
 
 | | Structural analysis of the Zn-form I of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1DC3
 
 | |
1LTV
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM PHENYLALANINE HYDROXYLASE, STRUCTURE WITH BOUND OXIDIZED Fe(III) | | Descriptor: | FE (III) ION, PHENYLALANINE-4-HYDROXYLASE | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
1ESM
 
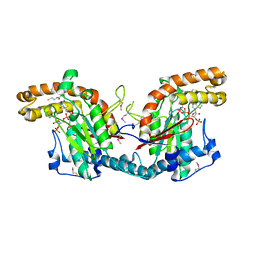 | | STRUCTURAL BASIS FOR THE FEEDBACK REGULATION OF ESCHERICHIA COLI PANTOTHENATE KINASE BY COENZYME A | | Descriptor: | COENZYME A, PANTOTHENATE KINASE | | Authors: | Yun, M, Park, C.G, Kim, J.Y, Rock, C.O, Jackowski, S, Park, H.W. | | Deposit date: | 2000-04-10 | | Release date: | 2000-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the feedback regulation of Escherichia coli pantothenate kinase by coenzyme A.
J.Biol.Chem., 275, 2000
|
|
1ESN
 
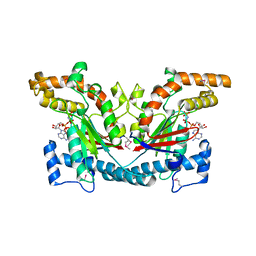 | | STRUCTURAL BASIS FOR THE FEEDBACK REGULATION OF ESCHERICHIA COLI PANTOTHENATE KINASE BY COENZYME A | | Descriptor: | MAGNESIUM ION, PANTOTHENATE KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yun, M, Park, C.G, Kim, J.Y, Rock, C.O, Jackowski, S, Park, H.W. | | Deposit date: | 2000-04-10 | | Release date: | 2000-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the feedback regulation of Escherichia coli pantothenate kinase by coenzyme A.
J.Biol.Chem., 275, 2000
|
|
1ZKJ
 
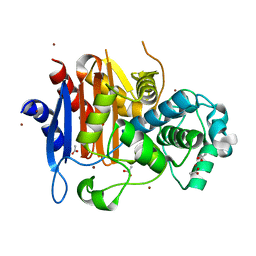 | | Structural Basis for the Extended Substrate Spectrum of CMY-10, a Plasmid-Encoded Class C beta-lactamase | | Descriptor: | ACETIC ACID, ZINC ION, extended-spectrum beta-lactamase | | Authors: | Cha, S.S, Jung, H.I, An, Y.J, Lee, S.H. | | Deposit date: | 2005-05-03 | | Release date: | 2006-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C beta-lactamase.
Mol.Microbiol., 60, 2006
|
|
5H8Y
 
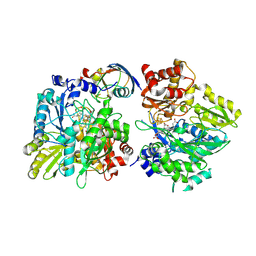 | | Crystal structure of the complex between maize sulfite reductase and ferredoxin in the form-2 crystal | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, ... | | Authors: | Kurisu, G, Nakayama, M, Hase, T. | | Deposit date: | 2015-12-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mutational studies of an electron transfer complex of maize sulfite reductase and ferredoxin.
J.Biochem., 160, 2016
|
|
