7RGU
 
 | |
7RGS
 
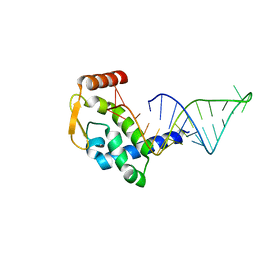
 | | The crystal structure of RocC, containing FinO domain, 24-126 | | Descriptor: | Repressor of competence, RNA Chaperone | | Authors: | Kim, H.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones.
Nat Commun, 13, 2022
|
|
7RGT
 
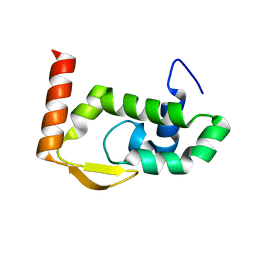
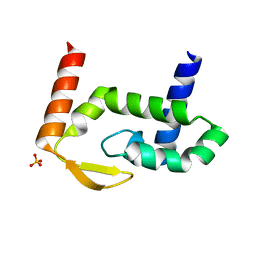 | | The crystal structure of RocC, containing FinO domain, 1-126 | | Descriptor: | Repressor of competence, RNA Chaperone, SULFATE ION | | Authors: | Kim, H.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones.
Nat Commun, 13, 2022
|
|
4Y1E
 
 | | SAV1875-C105D | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-07 | | Release date: | 2016-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
4Y0N
 
 | | SAV1875 | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-06 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
4Y1F
 
 | | SAV1875-E17D | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-07 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
4Y1R
 
 | | SAV1875-cysteinesulfonic acid | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-08 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
4Y1G
 
 | | SAV1875-E17N | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-07 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
3WUT
 
 | | Structure basis of inactivating cell abscission | | Descriptor: | Centrosomal protein of 55 kDa, GLYCEROL, Inactive serine/threonine-protein kinase TEX14 | | Authors: | Kim, H.J, Matsuura, A, Lee, H.H. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3WUV
 
 | | Structure basis of inactivating cell abscission with chimera peptide 2 | | Descriptor: | Centrosomal protein of 55 kDa, peptide from Programmed cell death 6-interacting protein | | Authors: | Kim, H.J, Matsuura, A, Lee, H.H. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3WUU
 
 | |
7B01
 
 | | ADAMTS13-CUB12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Maltodextrin-binding protein,Maltodextrin-binding protein,Maltodextrin-binding protein,ADAMTS13 CUB12,A disintegrin and metalloproteinase with thrombospondin motifs 13,A disintegrin and metalloproteinase with thrombospondin motifs 13,A disintegrin and metalloproteinase with thrombospondin motifs 13, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kim, H.J, Emsley, J. | | Deposit date: | 2020-11-18 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ADAMTS13 CUB domains reveals their role in global latency.
Sci Adv, 7, 2021
|
|
7CTO
 
 | | Staphylococcus aureus MsrB | | Descriptor: | Peptide methionine sulfoxide reductase MsrB | | Authors: | Kim, H.J. | | Deposit date: | 2020-08-19 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Implication of Staphylococcus aureus MsrB dimerization upon oxidation.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
5GNA
 
 | |
7W5R
 
 | | KRAS G12V and peptide complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, LEU-TYR-ASP-VAL-ALA, ... | | Authors: | Kim, H.J, Han, C.W, Jang, S.B. | | Deposit date: | 2021-11-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.87 Å) | | Cite: | Structural basis of the oncogenic KRAS mutant and GJ101 complex.
Biochem.Biophys.Res.Commun., 641, 2023
|
|
5XR2
 
 | | SAV0551 | | Descriptor: | LACTIC ACID, Protein/nucleic acid deglycase HchA, ZINC ION | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2017-06-07 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional studies of SAV0551 fromStaphylococcus aureusas a chaperone and glyoxalase III.
Biosci. Rep., 37, 2017
|
|
5XR3
 
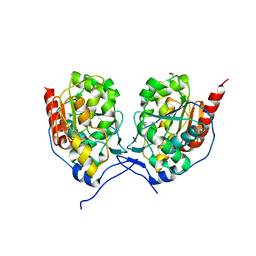 | | SAV0551 with glyoxylate | | Descriptor: | GLYOXYLIC ACID, Protein/nucleic acid deglycase HchA | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2017-06-08 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural and functional studies of SAV0551 fromStaphylococcus aureusas a chaperone and glyoxalase III.
Biosci. Rep., 37, 2017
|
|
5YU2
 
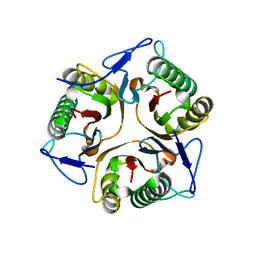 | | Structure of Ribonuclease YabJ | | Descriptor: | Translation initiation inhibitor homologue | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2017-11-20 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A novel chlorination-induced ribonuclease YabJ fromStaphylococcus aureus.
Biosci. Rep., 38, 2018
|
|
6KHM
 
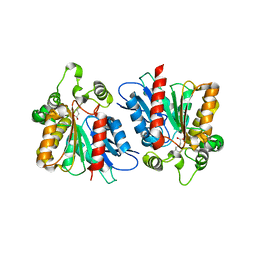 | | Lipase (Open form) | | Descriptor: | Hydrolase, alpha/beta domain protein, heptane-1,1-diol, ... | | Authors: | Kim, H.J, Kwon, A.R. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Closed, blocked, and open states of lipase from type II Cutibacterium acnes
To Be Published
|
|
6KHK
 
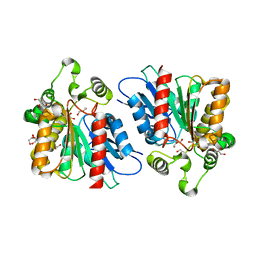 | | Lipase (Closed form) | | Descriptor: | GLYCEROL, Hydrolase, alpha/beta domain protein | | Authors: | Kim, H.J, Kwon, A.R. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Closed, blocked, and open states of lysophospholipase from type II Cutibacterium acnes
To Be Published
|
|
6KHL
 
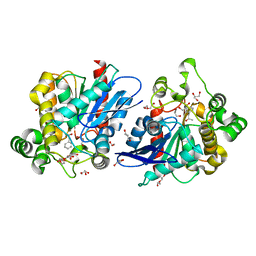 | | Lipase (Blocked form) | | Descriptor: | (2~{R})-1-phenylpropan-2-ol, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, H.J, Kwon, A.R. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Closed, blocked, and open states of lipase from type II Cutibacterium acnes
To Be Published
|
|
5D4Z
 
 | |
7EWC
 
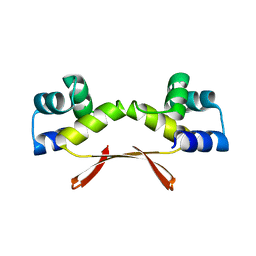 | | Mycobacterium tuberculosis HigA2 (Form I) | | Descriptor: | Putative antitoxin HigA2 | | Authors: | Kim, H.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Chasing the structural diversity of the transcription regulator Mycobacterium tuberculosis HigA2.
Iucrj, 8, 2021
|
|
7EWE
 
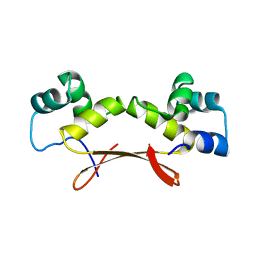 | | Mycobacterium tuberculosis HigA2 (Form III) | | Descriptor: | Putative antitoxin HigA2 | | Authors: | Kim, H.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Chasing the structural diversity of the transcription regulator Mycobacterium tuberculosis HigA2.
Iucrj, 8, 2021
|
|
7EWD
 
 | | Mycobacterium tuberculosis HigA2 (Form II) | | Descriptor: | Putative antitoxin HigA2 | | Authors: | Kim, H.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chasing the structural diversity of the transcription regulator Mycobacterium tuberculosis HigA2.
Iucrj, 8, 2021
|
|
