6JF6
 
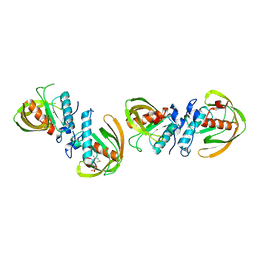 | |
6JES
 
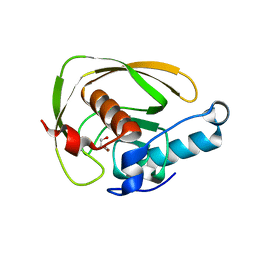 | |
6JF7
 
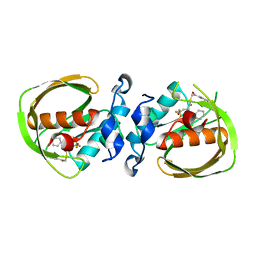 | | K3U bound crystal structure of class I type b peptide deformylase from Acinetobacter baumannii | | Descriptor: | Peptide deformylase, S-(2-oxo-2-phenylethyl) (2R)-2-benzyl-4,4,4-trifluorobutanethioate, ZINC ION | | Authors: | Jung, K.H, Ho, T.H, Lee, I.H, Kang, L.W. | | Deposit date: | 2019-02-08 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | K3U bound crystal structure of class I type b peptide deformylase from Acinetobacter baumannii
To be published
|
|
7D1I
 
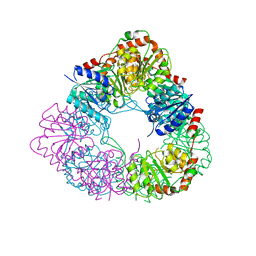 | | Crystal structure of acinetobacter baumannii MurG | | Descriptor: | UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase | | Authors: | Park, H.H, Jeong, k.H. | | Deposit date: | 2020-09-14 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.487 Å) | | Cite: | Putative hexameric glycosyltransferase functional unit revealed by the crystal structure of Acinetobacter baumannii MurG
Iucrj, 8, 2021
|
|
7D27
 
 | |
2M3G
 
 | | Structure of Anabaena Sensory Rhodopsin Determined by Solid State NMR Spectroscopy | | Descriptor: | Anabaena Sensory Rhodopsin, RETINAL | | Authors: | Wang, S, Munro, R.A, Shi, L, Kawamura, I, Okitsu, T, Wada, A, Kim, S, Jung, K, Brown, L.S, Ladizhansky, V. | | Deposit date: | 2013-01-17 | | Release date: | 2013-08-21 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein.
Nat.Methods, 10, 2013
|
|
5AWZ
 
 | | Crystal Structure of the Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, at 1.57 angstrom | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DECANE, DODECANE, ... | | Authors: | Furuse, M, Hosaka, T, Kimura-Someya, T, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5AX0
 
 | | Crystal Structure of the Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, at 1.52 angstrom | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DECANE, DODECANE, ... | | Authors: | Furuse, M, Hosaka, T, Kimura-Someya, T, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5AX1
 
 | | Crystal Structure of the Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, at 1.80 angstrom | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DECANE, DODECANE, ... | | Authors: | Furuse, M, Hosaka, T, Kimura-Someya, T, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7BXZ
 
 | |
6KU6
 
 | | OSM1 mutant - R326A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fumarate reductase 2, SUCCINIC ACID | | Authors: | Park, H.H, Kim, C.M. | | Deposit date: | 2019-08-30 | | Release date: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Crystal Structure of the Active Site Mutant Form of Soluble Fumarate Reductase, Osm1
Crystals, 2019
|
|
