5YDW
 
 | | Full-length structure of HypT from Salmonella typhimuriuma (hypochlorite-specific LysR-type transcriptional regulator) | | Descriptor: | Cell density-dependent motility repressor | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5YDV
 
 | | Regulatory domain of HypT from Salmonella typhimurium complexed with HOCl (HOCl-bound form) | | Descriptor: | Cell density-dependent motility repressor, SULFATE ION, hypochlorous acid | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-14 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5YEZ
 
 | | Regulatory domain of HypT M206Q mutant from Salmonella typhimurium | | Descriptor: | Cell density-dependent motility repressor | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-20 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5YER
 
 | | Regulatory domain of HypT from Salmonella typhimurium (Bromide ion-bound) | | Descriptor: | BROMIDE ION, Cell density-dependent motility repressor, SULFATE ION | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-19 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5YDO
 
 | | Regulatory domain of HypT from Salmonella typhimurium (apo-form) | | Descriptor: | Cell density-dependent motility repressor, SULFATE ION | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-13 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5X0Q
 
 | | OxyR2 E204G variant (Cl-bound) from Vibrio vulnificus | | Descriptor: | CHLORIDE ION, CITRIC ACID, LysR family transcriptional regulator | | Authors: | Jo, I, Ha, N.-C. | | Deposit date: | 2017-01-23 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The hydrogen peroxide hypersensitivity of OxyR2 in Vibrio vulnificus depends on conformational constraints
J. Biol. Chem., 292, 2017
|
|
5X0V
 
 | | Reduced form of regulatory domain of OxyR2 from Vibrio vulnificus | | Descriptor: | CHLORIDE ION, CITRIC ACID, LysR family transcriptional regulator | | Authors: | Jo, I, Ha, N.-C. | | Deposit date: | 2017-01-23 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The hydrogen peroxide hypersensitivity of OxyR2 in Vibrio vulnificus depends on conformational constraints
J. Biol. Chem., 292, 2017
|
|
5HFG
 
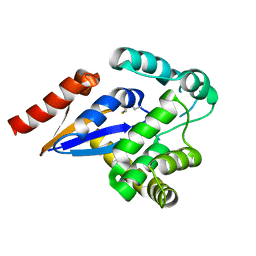 | | Cytosolic disulfide reductase DsbM from Pseudomonas aeruginosa | | Descriptor: | Uncharacterized protein, cytosolic disulfide reductase DsbM | | Authors: | Jo, I, Ha, N.-C. | | Deposit date: | 2016-01-07 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Crystal structures of the disulfide reductase DsbM from Pseudomonas aeruginosa
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5HFI
 
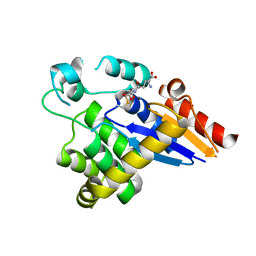 | |
5B7H
 
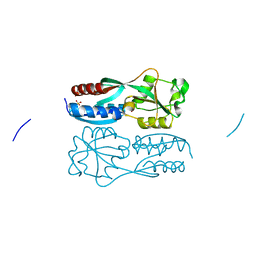 | |
5B7D
 
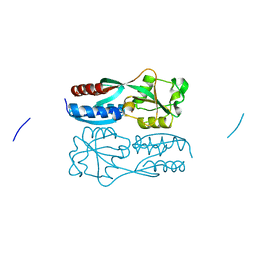 | |
5B70
 
 | | OxyR2 E204G regulatory domain from Vibrio vulnificus | | Descriptor: | GLYCEROL, LysR family transcriptional regulator | | Authors: | Jo, I, Ha, N.-C. | | Deposit date: | 2016-06-02 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The hydrogen peroxide hypersensitivity of OxyR2 in Vibrio vulnificus depends on conformational constraints
J. Biol. Chem., 292, 2017
|
|
4X6G
 
 | |
4Y0M
 
 | |
4XWS
 
 | |
7X5D
 
 | |
5Y9S
 
 | | Crystal structure of VV2_1132, a LysR family transcriptional regulator | | Descriptor: | BROMIDE ION, VV2_1132 | | Authors: | Jang, Y, Hong, S, Jo, I, Ahn, J, Ha, N.C. | | Deposit date: | 2017-08-28 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | A Novel Tetrameric Assembly Configuration in VV2_1132, a LysR-Type Transcriptional Regulator inVibrio vulnificus
Mol. Cells, 41, 2018
|
|
5ZQJ
 
 | | Crystal structure of beta-xylosidase from Bacillus pumilus | | Descriptor: | Beta-xylosidase, GLYCEROL | | Authors: | Ha, N.C, Hong, S, Jo, I. | | Deposit date: | 2018-04-19 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-based protein engineering of bacterial beta-xylosidase to increase the production yield of xylobiose from xylose
Biochem. Biophys. Res. Commun., 501, 2018
|
|
5ZQS
 
 | |
5ZQX
 
 | |
5X15
 
 | |
4PWO
 
 | | Crystal structure of DsbA from the Gram positive bacterium Corynebacterium diphtheriae | | Descriptor: | DsbA, GLYCEROL | | Authors: | Um, S.H, Kim, J.S, Jiao, L, Yoon, B.Y, Jo, I, Ha, N.C. | | Deposit date: | 2014-03-21 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure and biochemical characterization of DsbA from the Gram positive bacterium Corynebacterium diphtheriae
To be Published
|
|
4PWP
 
 | | Crystal structure of DsbA from the Gram positive bacterium Corynebacterium diphtheriae | | Descriptor: | DsbA, GLYCEROL | | Authors: | Um, S.H, Kim, J.S, Jiao, L, Yoon, B.Y, Jo, I, Ha, N.C. | | Deposit date: | 2014-03-21 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Crystal structure and biochemical characterization of DsbA from the Gram positive bacterium Corynebacterium diphtheriae
To be Published
|
|
5WU7
 
 | |
6JLB
 
 | |
