2P1J
 
 | | Crystal structure of a polC-type DNA polymerase III exonuclease domain from Thermotoga maritima | | Descriptor: | DNA polymerase III polC-type | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Izuka, M, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-05 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a polC-type DNA polymerase III exonuclease domain from Thermotoga maritima
To be Published
|
|
5XOF
 
 | | Crystal structure of human paired immunoglobulin-like type 2 receptor alpha with synthesized glycopeptide I | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-6)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Paired immunoglobulin-like type 2 receptor alpha, Peptide from Nitric oxide synthase, ... | | Authors: | Furukawa, A, Kakita, K, Yamada, T, Ishizuka, M, Sakamoto, J, Hatori, N, Maeda, N, Ohsaka, F, Saitoh, T, Nomura, T, Kuroki, K, Nambu, H, Arase, H, Matsunaga, H, Anada, M, Ose, T, Hashimoto, S, Maenaka, K. | | Deposit date: | 2017-05-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.963 Å) | | Cite: | Structural and thermodynamic analyses reveal critical features of glycopeptide recognition by the human PILR alpha immune cell receptor.
J. Biol. Chem., 292, 2017
|
|
3MGK
 
 | | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum ATCC 824 | | Descriptor: | Intracellular protease/amidase related enzyme (ThiJ family) | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-06 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum
To be Published
|
|
3GT5
 
 | | Crystal structure of an N-acetylglucosamine 2-epimerase family protein from Xylella fastidiosa | | Descriptor: | CHLORIDE ION, N-acetylglucosamine 2-epimerase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an N-acetylglucosamine 2-epimerase family protein from Xylella fastidiosa
To be Published
|
|
3H5I
 
 | | Crystal structure of the N-terminal domain of a response regulator/sensory box/GGDEF 3-domain protein from Carboxydothermus hydrogenoformans | | Descriptor: | CHLORIDE ION, Response regulator/sensory box protein/GGDEF domain protein, SODIUM ION | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Iizuka, M, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the N-terminal domain of a response regulator/sensory box/GGDEF 3-domain protein from Carboxydothermus hydrogenoformans
To be Published
|
|
3HJG
 
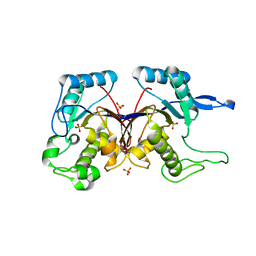 | | Crystal structure of putative alpha-ribazole-5'-phosphate phosphatase CobC FROM Vibrio parahaemolyticus | | Descriptor: | Putative alpha-ribazole-5'-phosphate phosphatase CobC, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Rutter, M, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of putative alpha-ribazole-5' -phosphate phosphatase FROM Vibrio parahaemolyticus
To be Published
|
|
3G7U
 
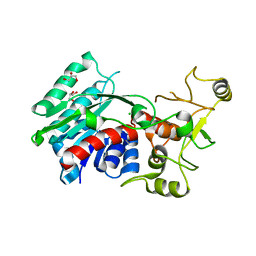 | | Crystal structure of putative DNA modification methyltransferase encoded within prophage Cp-933R (E.coli) | | Descriptor: | CHLORIDE ION, Cytosine-specific methyltransferase, GLYCEROL | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Gilmore, M, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-10 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of DNA Modification Methyltransferase Encoded within Prophage Cp-933R (E.coli)
To be Published
|
|
3GM8
 
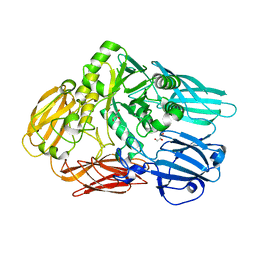 | | Crystal structure of a beta-glycosidase from Bacteroides vulgatus | | Descriptor: | GLYCEROL, Glycoside hydrolase family 2, candidate beta-glycosidase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-13 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a beta-glycosidase from Bacteroides vulgatus
To be Published
|
|
3BY5
 
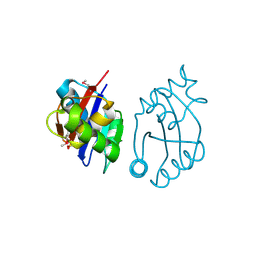 | | Crystal structure of cobalamin biosynthesis protein chiG from Agrobacterium tumefaciens str. C58 | | Descriptor: | Cobalamin biosynthesis protein, SULFATE ION | | Authors: | Patskovsky, Y, Bonanno, J.B, Sojitra, S, Rutter, M, Iizuka, M, Maletic, M, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of cobalamin biosynthesis protein from Agrobacterium tumefaciens str. C58.
To be Published
|
|
3BVC
 
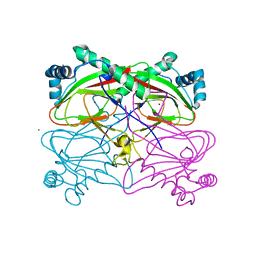 | | Crystal structure of uncharacterized protein Ism_01780 from Roseovarius nubinhibens ISM | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Uncharacterized protein Ism_01780 | | Authors: | Patskovsky, Y, Toro, R, Meyer, A.J, Rutter, M, Iizuka, M, Maletic, M, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-06 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of an uncharacterized protein Ism_01780 from Roseovarius nubinhibens.
To be Published
|
|
2QH5
 
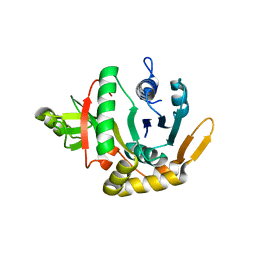 | | Crystal structure of mannose-6-phosphate isomerase from Helicobacter pylori | | Descriptor: | Mannose-6-phosphate isomerase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Sauder, J.M, Dickey, M, Iizuka, M, Maletic, M, Wasserman, S.R, Koss, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mannose-6-phosphate isomerase from Helicobacter pylori.
To be Published
|
|
3BMA
 
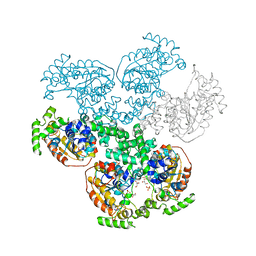 | | Crystal structure of D-alanyl-lipoteichoic acid synthetase from Streptococcus pneumoniae R6 | | Descriptor: | D-alanyl-lipoteichoic acid synthetase, GLYCEROL, SULFATE ION | | Authors: | Patskovsky, Y, Sridhar, V, Bonanno, J.B, Smith, D, Rutter, M, Iizuka, M, Koss, J, Bain, K, Gheyi, T, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-12 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of probable D-Alanyl-Lipoteichoic Acid Synthetase from Streptococcus pneumoniae.
To be Published
|
|
3CBW
 
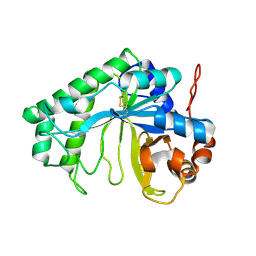 | | Crystal structure of the YdhT protein from Bacillus subtilis | | Descriptor: | CITRIC ACID, YdhT protein | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-23 | | Release date: | 2008-03-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.269 Å) | | Cite: | Crystal structure of the YdhT protein from Bacillus subtilis.
To be Published
|
|
3CDX
 
 | | Crystal structure of succinylglutamatedesuccinylase/aspartoacylase from Rhodobacter sphaeroides | | Descriptor: | CALCIUM ION, Succinylglutamatedesuccinylase/aspartoacylase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Patterson, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-27 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of succinylglutamatedesuccinylase/aspartoacylase from Rhodobacter sphaeroides.
To be Published
|
|
3DO9
 
 | | Crystal structure of protein ba1542 from bacillus anthracis str.ames | | Descriptor: | UPF0302 protein BA_1542/GBAA1542/BAS1430 | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Iizuka, M, Maletic, M, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-03 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Protein Ba1542 from Bacillus Anthracis Str.Ames.
To be Published
|
|
3ERV
 
 | | Crystal structure of an putative C39-like peptidase from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, putative C39-like peptidase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-03 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an putative C39-like peptidase from Bacillus anthracis
To be Published
|
|
3EQZ
 
 | | Crystal structure of a response regulator from Colwellia psychrerythraea | | Descriptor: | Response regulator | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Iizuka, M, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of a response regulator from Colwellia psychrerythraea
To be Published
|
|
3DHU
 
 | | Crystal structure of an alpha-amylase from Lactobacillus plantarum | | Descriptor: | Alpha-amylase | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-18 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an alpha-amylase from Lactobacillus plantarum
To be Published
|
|
3DLS
 
 | | Crystal structure of human PAS kinase bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PAS domain-containing serine/threonine-protein kinase | | Authors: | Antonysamy, S, Bonanno, J.B, Romero, R, Russell, M, Iizuka, M, Gheyi, T, Wasserman, S.R, Rutter, J, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-29 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural bases of PAS domain-regulated kinase (PASK) activation in the absence of activation loop phosphorylation.
J.Biol.Chem., 285, 2010
|
|
2QV5
 
 | | Crystal structure of uncharacterized protein ATU2773 from Agrobacterium tumefaciens C58 | | Descriptor: | GLYCEROL, Uncharacterized protein Atu2773 | | Authors: | Patskovsky, Y, Bonanno, J.B, Sojitra, S, Dickey, M, Bain, K.T, Iizuka, M, Smith, D, Rodgers, L, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-07 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Uncharacterized Protein Atu2773 from Agrobacterium Tumefaciens.
To be Published
|
|
5XO2
 
 | | Crystal structure of human paired immunoglobulin-like type 2 receptor alpha with synthesized glycopeptide II | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-6)-2-acetamido-2,4-dideoxy-alpha-D-xylo-hexopyranose, Paired immunoglobulin-like type 2 receptor alpha, Peptide from Envelope glycoprotein B | | Authors: | Furukawa, A, Kakita, K, Yamada, T, Ishizuka, M, Sakamoto, J, Hatori, N, Maeda, N, Ohsaka, F, Saitoh, T, Nomura, T, Kuroki, K, Nambu, H, Arase, H, Matsunaga, S, Anada, M, Ose, T, Hashimoto, S, Maenaka, K. | | Deposit date: | 2017-05-25 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and thermodynamic analyses reveal critical features of glycopeptide recognition by the human PILR alpha immune cell receptor.
J. Biol. Chem., 292, 2017
|
|
2PTF
 
 | | Crystal structure of protein MTH_863 from Methanobacterium thermoautotrophicum bound to FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Uncharacterized protein MTH_863 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-08 | | Release date: | 2007-05-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of protein MTH_863 from Methanobacterium thermoautotrophicum bound to FMN.
To be Published
|
|
2PMB
 
 | | Crystal structure of predicted nucleotide-binding protein from Vibrio cholerae | | Descriptor: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein | | Authors: | Patskovsky, Y, Zhan, C, Shi, W, Toro, R, Sauder, J.M, Gilmore, J, Iizuka, M, Maletic, M, Gheyi, T, Wasserman, S.R, Smith, D, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of Predicted Nucleotide-Binding Protein from Vibrio Cholerae.
To be Published
|
|
2QSR
 
 | | Crystal structure of C-terminal domain of transcription-repair coupling factor | | Descriptor: | Transcription-repair coupling factor | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Bain, K, Iizuka, M, Wasserman, S, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-31 | | Release date: | 2007-09-11 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of C-terminal domain of transcription-repair coupling factor.
To be Published
|
|
2PNW
 
 | | Crystal structure of membrane-bound lytic murein transglycosylase from Agrobacterium tumefaciens | | Descriptor: | Membrane-bound lytic murein transglycosylase, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-25 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of membrane-bound lytic murein transglycosylase from Agrobacterium tumefaciens.
To be Published
|
|
