4UWM
 
 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2014-08-12 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5AEC
 
 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2015-08-28 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ERG
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, FE (III) ION | | Authors: | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | Deposit date: | 2012-04-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
4EPK
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | Authors: | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | Deposit date: | 2012-04-17 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6009 Å) | | Cite: | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
4ERI
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | Authors: | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | Deposit date: | 2012-04-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0006 Å) | | Cite: | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
4ERA
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | Deposit date: | 2012-04-19 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
4OFC
 
 | | 2.0 Angstroms X-ray crystal structure of human 2-amino-3-carboxymuconate-6-semialdehye decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase, ZINC ION | | Authors: | Huo, L, Liu, F, Iwaki, H, Chen, L, Hasegawa, Y, Liu, A. | | Deposit date: | 2014-01-14 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Human alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase (ACMSD): A structural and mechanistic unveiling.
Proteins, 83, 2015
|
|
4OE2
 
 | | 2.00 Angstroms X-ray crystal structure of E268A 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Davis, I, Liu, F, Esaki, S, Iwaki, H, Hasegawa, Y, Liu, A. | | Deposit date: | 2014-01-11 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4NPI
 
 | | 1.94 Angstroms X-ray crystal structure of NAD- and intermediate- bound alpha-aminomuconate-epsilon-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | (2Z,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Davis, I, Liu, F, Iwaki, H, Hasegawa, Y, Liu, A. | | Deposit date: | 2013-11-21 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
8GST
 
 | | Crystal structure of L-2,4-diketo-3-deoxyrhamnonate hydrolase from Sphingomonas sp. (pyruvate bound-form) | | Descriptor: | L-2,4-diketo-3-deoxyrhamnonate hydrolase, MAGNESIUM ION, PYRUVIC ACID | | Authors: | Fukuhara, S, Watanabe, Y, Watanabe, S, Nishiwaki, H. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structure of l-2,4-Diketo-3-deoxyrhamnonate Hydrolase Involved in the Nonphosphorylated l-Rhamnose Pathway from Bacteria.
Biochemistry, 62, 2023
|
|
8GSR
 
 | | Crystal structure of L-2,4-diketo-3-deoxyrhamnonate hydrolase from Sphingomonas sp. (apo-form) | | Descriptor: | L-2,4-diketo-3-deoxyrhamnonate hydrolase, MAGNESIUM ION | | Authors: | Fukuhara, S, Watanabe, Y, Watanabe, S, Nishiwaki, H. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of l-2,4-Diketo-3-deoxyrhamnonate Hydrolase Involved in the Nonphosphorylated l-Rhamnose Pathway from Bacteria.
Biochemistry, 62, 2023
|
|
3GWF
 
 | | Open crystal structure of cyclohexanone monooxygenase | | Descriptor: | Cyclohexanone monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mirza, I.A, Yachnin, B.J, Berghuis, A.M. | | Deposit date: | 2009-04-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of cyclohexanone monooxygenase reveal complex domain movements and a sliding cofactor
J.Am.Chem.Soc., 131, 2009
|
|
3GWD
 
 | | Closed crystal structure of cyclohexanone monooxygenase | | Descriptor: | Cyclohexanone monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mirza, I.A, Yachnin, B.J, Berghuis, A.M. | | Deposit date: | 2009-03-31 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of cyclohexanone monooxygenase reveal complex domain movements and a sliding cofactor
J.Am.Chem.Soc., 131, 2009
|
|
3UOX
 
 | | Crystal Structure of OTEMO (FAD bound form 2) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UOY
 
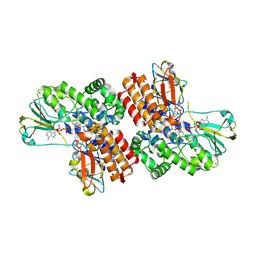 | | Crystal Structure of OTEMO complex with FAD and NADP (form 1) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO, ... | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UOV
 
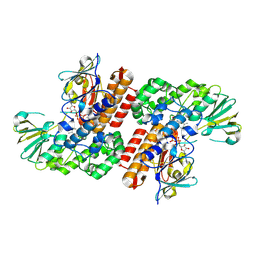 | | Crystal Structure of OTEMO (FAD bound form 1) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UP5
 
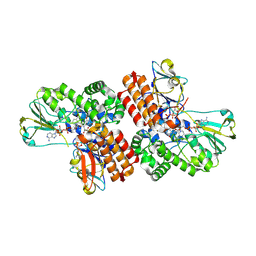 | | Crystal Structure of OTEMO complex with FAD and NADP (form 4) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UP4
 
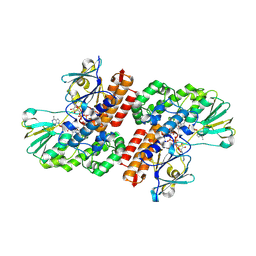 | | Crystal Structure of OTEMO complex with FAD and NADP (form 3) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UOZ
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 2) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
4OUB
 
 | | A 2.20 angstroms X-ray crystal structure of E268A 2-aminomucaonate 6-semialdehyde dehydrogenase catalytic intermediate from Pseudomonas fluorescens | | Descriptor: | (2Z,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Liu, A. | | Deposit date: | 2014-02-15 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4OU2
 
 | | A 2.15 Angstroms X-ray crystal structure of E268A 2-aminomuconate 6-semialdehyde dehydrogenase catalytic intermediate from Pseudomonas fluorescens | | Descriptor: | (2Z,4E)-2,6-dihydroxyhexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Liu, A. | | Deposit date: | 2014-02-14 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4I59
 
 | |
4I25
 
 | | 2.00 Angstroms X-ray crystal structure of NAD- and substrate-bound 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | (2E,4E)-2-amino-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Davis, I, Chen, L, Liu, A. | | Deposit date: | 2012-11-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4I2R
 
 | | 2.15 Angstroms X-ray crystal structure of NAD- and alternative substrate-bound 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | (2E,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Davis, I, Chen, L, Liu, A. | | Deposit date: | 2012-11-22 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4I58
 
 | |
