1IQP
 
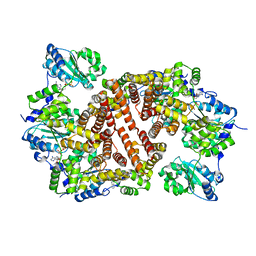 | | Crystal Structure of the Clamp Loader Small Subunit from Pyrococcus furiosus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RFCS | | Authors: | Oyama, T, Ishino, Y, Cann, I.K.O, Ishino, S, Morikawa, K. | | Deposit date: | 2001-07-24 | | Release date: | 2001-09-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic Structure of the Clamp Loader Small Subunit from Pyrococcus furiosus
Mol.Cell, 8, 2001
|
|
7E15
 
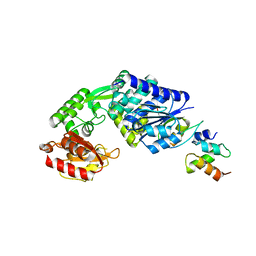 | |
1WP9
 
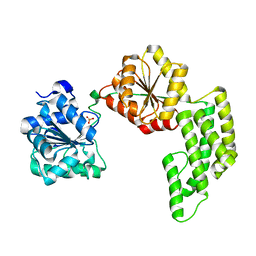 | | Crystal structure of Pyrococcus furiosus Hef helicase domain | | Descriptor: | ATP-dependent RNA helicase, putative, PHOSPHATE ION | | Authors: | Nishino, T, Komori, K, Tsuchiya, D, Ishino, Y, Morikawa, K. | | Deposit date: | 2004-08-31 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure and Functional Implications of Pyrococcus furiosus Hef Helicase Domain Involved in Branched DNA Processing
Structure, 13, 2005
|
|
1X2I
 
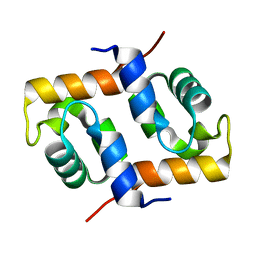 | | Crystal Structure Of Archaeal Xpf/Mus81 Homolog, Hef From Pyrococcus Furiosus, Helix-hairpin-helix Domain | | Descriptor: | Hef helicase/nuclease | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2005-04-24 | | Release date: | 2005-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Analyses of an Archaeal XPF/Rad1/Mus81 Nuclease: Asymmetric DNA Binding and Cleavage Mechanisms
STRUCTURE, 13, 2005
|
|
6JI2
 
 | | Crystal structure of archaeal ribosomal protein aP1, aPelota, and GTP-bound aEF1A complex | | Descriptor: | Archaeal ribosomal stalk protein aP1, Elongation factor 1-alpha, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Maruyama, K, Imai, H, Kawamura, M, Ishino, S, Ishino, Y, Ito, K, Uchiumi, T. | | Deposit date: | 2019-02-20 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Switch of the interactions between the ribosomal stalk and EF1A in the GTP- and GDP-bound conformations.
Sci Rep, 9, 2019
|
|
6KNC
 
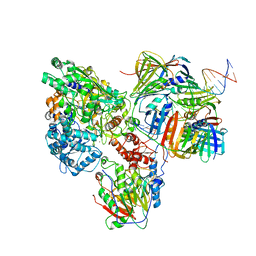 | | PolD-PCNA-DNA (form B) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
6KNB
 
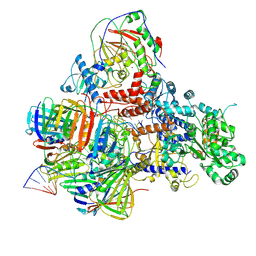 | | PolD-PCNA-DNA (form A) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
1IZ4
 
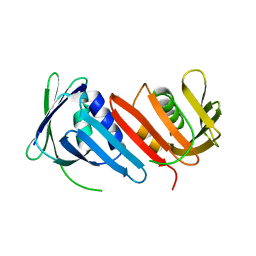 | |
1ISQ
 
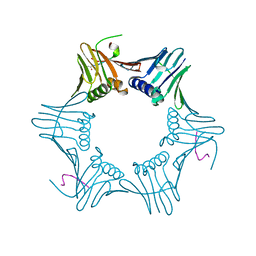 | | Pyrococcus furiosus PCNA complexed with RFCL PIP-box peptide | | Descriptor: | Proliferating Cell Nuclear Antigen, replication factor C large subunit | | Authors: | Matsumiya, S, Ishino, S, Ishino, Y, Morikawa, K. | | Deposit date: | 2001-12-19 | | Release date: | 2002-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Physical interaction between proliferating cell nuclear antigen and
replication factor C from Pyrococcus furiosus
Genes Cells, 7, 2002
|
|
1IZ5
 
 | | Pyrococcus furiosus PCNA mutant (Met73Leu, Asp143Ala, Asp147Ala): orthorhombic form | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Matsumiya, S, Ishino, S, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-09-23 | | Release date: | 2003-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Intermolecular ion pairs maintain the toroidal structure of Pyrococcus furiosus PCNA
PROTEIN SCI., 12, 2003
|
|
5GKI
 
 | | Structure of EndoMS-dsDNA3 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*CP*CP*TP*AP*GP*GP*TP*CP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*GP*GP*GP*CP*CP*TP*AP*GP*GP*C)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKG
 
 | | Structure of EndoMS-dsDNA1'' complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*CP*GP*CP*TP*AP*CP*AP*GP*GP*TP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*CP*GP*TP*GP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKJ
 
 | | Structure of EndoMS in apo form | | Descriptor: | Endonuclease EndoMS | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKF
 
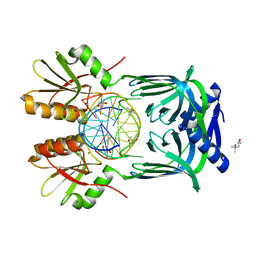 | | Structure of EndoMS-dsDNA1' complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*CP*GP*CP*TP*AP*CP*AP*TP*GP*TP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*CP*TP*TP*GP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKH
 
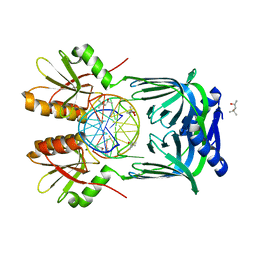 | | Structure of EndoMS-dsDNA2 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*CP*GP*GP*CP*AP*CP*TP*TP*GP*GP*CP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*GP*CP*CP*AP*GP*GP*TP*GP*CP*CP*GP*T)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKE
 
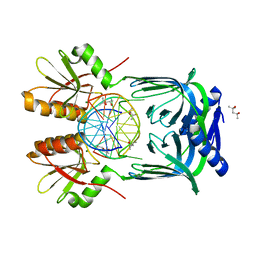 | | Structure of EndoMS-dsDNA1 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*CP*GP*CP*TP*AP*CP*AP*TP*GP*TP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*CP*GP*TP*GP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
1GEF
 
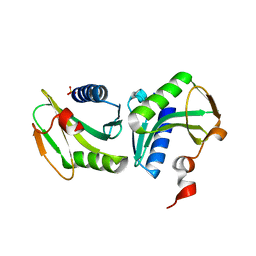 | | Crystal structure of the archaeal holliday junction resolvase HJC | | Descriptor: | HOLLIDAY JUNCTION RESOLVASE, SULFATE ION | | Authors: | Nishino, T, Komori, K, Tsuchiya, D, Ishino, Y, Morikawa, K. | | Deposit date: | 2000-11-08 | | Release date: | 2001-03-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the archaeal holliday junction resolvase Hjc and implications for DNA recognition.
Structure, 9, 2001
|
|
1IPI
 
 | |
1DQ3
 
 | |
1GE8
 
 | |
1J23
 
 | | Crystal structure of archaeal XPF/Mus81 homolog, Hef from Pyrococcus furiosus, nuclease domain | | Descriptor: | ATP-dependent RNA helicase, putative | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-12-25 | | Release date: | 2003-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-Ray and Biochemical Anatomy of an Archaeal XPF/Rad1/Mus81 Family Nuclease. Similarity between Its Endonuclease Domain and Restriction Enzymes
Structure, 11, 2003
|
|
1J25
 
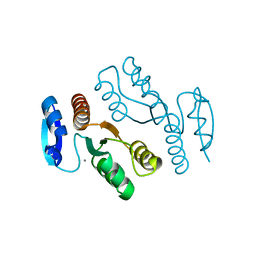 | | Crystal structure of archaeal XPF/Mus81 homolog, Hef from Pyrococcus furiosus, nuclease domain, Mn cocrystal | | Descriptor: | ATP-dependent RNA helicase, putative, MANGANESE (II) ION | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-12-25 | | Release date: | 2003-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-Ray and Biochemical Anatomy of an Archaeal XPF/Rad1/Mus81 Family Nuclease. Similarity between Its Endonuclease Domain and Restriction Enzymes
Structure, 11, 2003
|
|
1J24
 
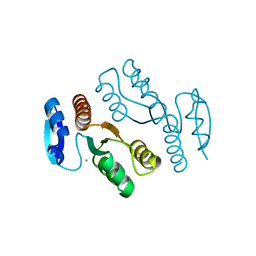 | | Crystal structure of archaeal XPF/Mus81 homolog, Hef from Pyrococcus furiosus, nuclease domain, Ca cocrystal | | Descriptor: | ATP-dependent RNA helicase, putative, CALCIUM ION | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-12-25 | | Release date: | 2003-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-Ray and Biochemical Anatomy of an Archaeal XPF/Rad1/Mus81 Family Nuclease. Similarity between Its Endonuclease Domain and Restriction Enzymes
Structure, 11, 2003
|
|
1J22
 
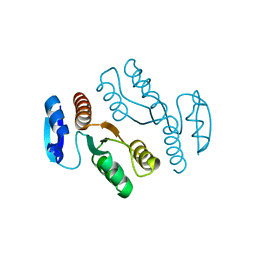 | | Crystal structure of archaeal XPF/Mus81 homolog, Hef from Pyrococcus furiosus, nuclease domain, selenomet derivative | | Descriptor: | ATP-dependent RNA helicase, putative | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-12-25 | | Release date: | 2003-04-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray and Biochemical Anatomy of an Archaeal XPF/Rad1/Mus81 Family Nuclease. Similarity between Its Endonuclease Domain and Restriction Enzymes
Structure, 11, 2003
|
|
2CFM
 
 | |
