2HI7
 
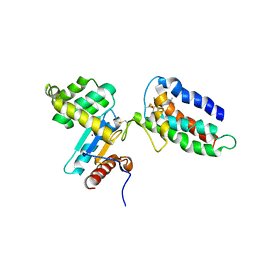 | | Crystal structure of DsbA-DsbB-ubiquinone complex | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein dsbA, UBIQUINONE-1, ... | | Authors: | Inaba, K, Murakami, S, Suzuki, M, Nakagawa, A, Yamashita, E, Okada, K, Ito, K. | | Deposit date: | 2006-06-29 | | Release date: | 2006-12-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of the DsbB-DsbA Complex Reveals a Mechanism of Disulfide Bond Generation
Cell(Cambridge,Mass.), 127, 2006
|
|
7EBT
 
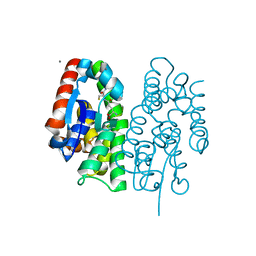 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in glutathione-bound form | | Descriptor: | CALCIUM ION, GLUTATHIONE, Glutathione transferase | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
7EBW
 
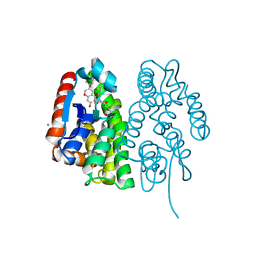 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in desmethylglycitein and glutathione-bound form | | Descriptor: | 6,7-dihydroxy-3-(4-hydroxyphenyl)-4H-chromen-4-one, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
7EBV
 
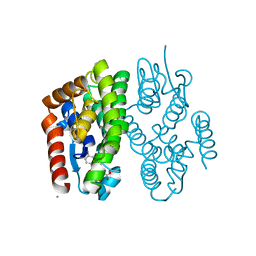 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in luteolin- and glutathione-bound form | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
7EBU
 
 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in Daidzein- and glutathione-bound form | | Descriptor: | 7-hydroxy-3-(4-hydroxyphenyl)-4H-chromen-4-one, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
2ZPL
 
 | | Crystal structure analysis of PDZ domain A | | Descriptor: | GLYCEROL, NICKEL (II) ION, Regulator of sigma E protease | | Authors: | Inaba, K, Suzuki, M. | | Deposit date: | 2008-07-17 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure analysis of PDZ-domain A
To be Published
|
|
2ZPM
 
 | |
2ZUQ
 
 | | Crystal structure of DsbB-Fab complex | | Descriptor: | Disulfide bond formation protein B, Fab fragment heavy chain, Fab fragment light chain, ... | | Authors: | Inaba, K, Suzuki, M, Murakami, S. | | Deposit date: | 2008-10-28 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dynamic nature of disulphide bond formation catalysts revealed by crystal structures of DsbB
Embo J., 28, 2009
|
|
2ZUP
 
 | | Updated crystal structure of DsbB-DsbA complex from E. coli | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein dsbA, UBIQUINONE-1, ... | | Authors: | Inaba, K, Suzuki, M, Murakami, S, Nakagawa, A. | | Deposit date: | 2008-10-28 | | Release date: | 2009-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Dynamic nature of disulphide bond formation catalysts revealed by crystal structures of DsbB
Embo J., 28, 2009
|
|
3AHR
 
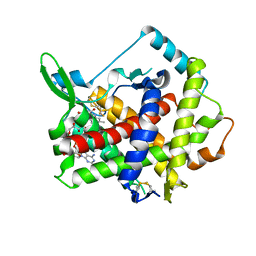 | | Inactive human Ero1 | | Descriptor: | ERO1-like protein alpha, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Inaba, K, Sitia, R, Suzuki, M. | | Deposit date: | 2010-04-26 | | Release date: | 2010-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Crystal structures of human Ero1-alpha reveal the mechanisms of regulated and targeted oxidation of PDI
Embo J., 29, 2010
|
|
3AHQ
 
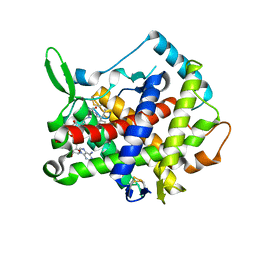 | | hyperactive human Ero1 | | Descriptor: | ERO1-like protein alpha, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Inaba, K, Sitia, R, Suzuki, M. | | Deposit date: | 2010-04-26 | | Release date: | 2010-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of human Ero1-alpha reveal the mechanisms of regulated and targeted oxidation of PDI
Embo J., 29, 2010
|
|
3WGD
 
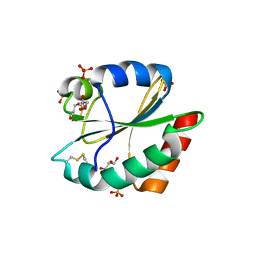 | | Crystal structure of ERp46 Trx1 | | Descriptor: | GLYCEROL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Inaba, K, Suzuki, M, Kojima, R. | | Deposit date: | 2013-08-04 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Radically different thioredoxin domain arrangement of ERp46, an efficient disulfide bond introducer of the mammalian PDI family
Structure, 22, 2014
|
|
3WGX
 
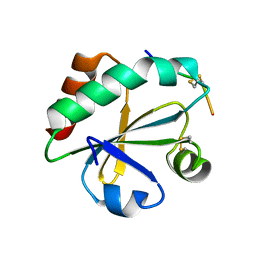 | | Crystal structure of ERp46 Trx2 in a complex with Prx4 C-term | | Descriptor: | GLYCEROL, Peroxiredoxin-4, Thioredoxin domain-containing protein 5 | | Authors: | Inaba, K, Suzuki, M, Kojima, R. | | Deposit date: | 2013-08-13 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Radically different thioredoxin domain arrangement of ERp46, an efficient disulfide bond introducer of the mammalian PDI family
Structure, 22, 2014
|
|
3APQ
 
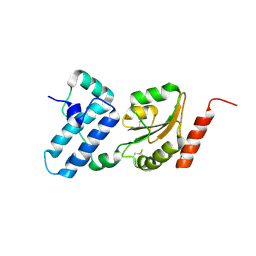 | |
3APO
 
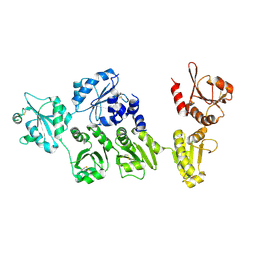 | | Crystal structure of full-length ERdj5 | | Descriptor: | DnaJ homolog subfamily C member 10 | | Authors: | Inaba, K, Suzuki, M, Nagata, K. | | Deposit date: | 2010-10-20 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5.
Mol.Cell, 41, 2011
|
|
3APS
 
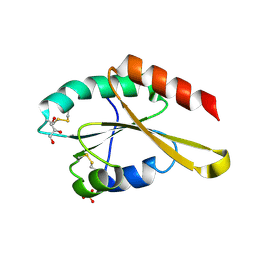 | | Crystal structure of Trx4 domain of ERdj5 | | Descriptor: | DnaJ homolog subfamily C member 10, GLYCEROL, SULFATE ION | | Authors: | Inaba, K, Suzuki, M, Nagata, K. | | Deposit date: | 2010-10-20 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5.
Mol.Cell, 41, 2011
|
|
3VWU
 
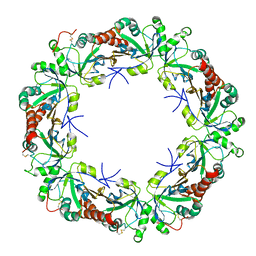 | |
3VWW
 
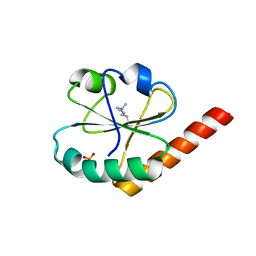 | | Crystal structure of a0-domain of P5 from H. sapiens | | Descriptor: | PHOSPHATE ION, Protein disulfide-isomerase A6 | | Authors: | Inaba, K, Suzuki, M. | | Deposit date: | 2012-09-03 | | Release date: | 2013-09-04 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Synergistic cooperation of PDI family members in peroxiredoxin 4-driven oxidative protein folding
Sci Rep, 3, 2013
|
|
3VWV
 
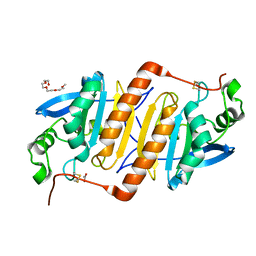 | |
3W8J
 
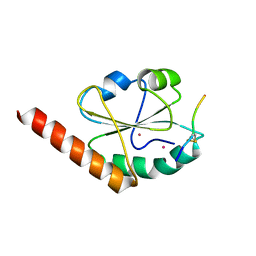 | | Crystal structure of P5 a0 in a complex with Prx4 c-term | | Descriptor: | C-terminal peptide from Peroxiredoxin-4, GLYCEROL, POTASSIUM ION, ... | | Authors: | Inaba, K, Suzuki, M, Kojima, R. | | Deposit date: | 2013-03-13 | | Release date: | 2013-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synergistic cooperation of PDI family members in peroxiredoxin 4-driven oxidative protein folding
Sci Rep, 3, 2013
|
|
3WGE
 
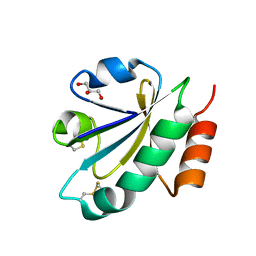 | | Crystal structure of ERp46 Trx2 | | Descriptor: | GLYCEROL, Thioredoxin domain-containing protein 5 | | Authors: | Inaba, K, Suzuki, M, Kojima, R. | | Deposit date: | 2013-08-04 | | Release date: | 2014-06-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Radically different thioredoxin domain arrangement of ERp46, an efficient disulfide bond introducer of the mammalian PDI family
Structure, 22, 2014
|
|
1CH4
 
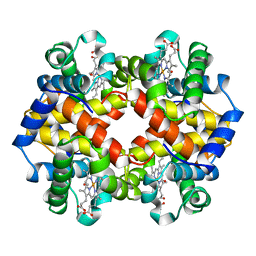 | | MODULE-SUBSTITUTED CHIMERA HEMOGLOBIN BETA-ALPHA (F133V) | | Descriptor: | CARBON MONOXIDE, MODULE-SUBSTITUTED CHIMERA HEMOGLOBIN BETA-ALPHA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shirai, T, Fujikake, M, Yamane, T, Inaba, K, Ishimori, K, Morishima, I. | | Deposit date: | 1998-06-11 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a protein with an artificial exon-shuffling, module M4-substituted chimera hemoglobin beta alpha, at 2.5 A resolution.
J.Mol.Biol., 287, 1999
|
|
5XWM
 
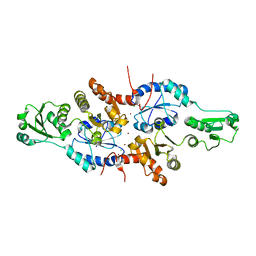 | | human ERp44 zinc-bound form | | Descriptor: | CHLORIDE ION, Endoplasmic reticulum resident protein 44, ZINC ION | | Authors: | Watanabe, S, Harayama, M, Inaba, K. | | Deposit date: | 2017-06-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Zinc regulates ERp44-dependent protein quality control in the early secretory pathway.
Nat Commun, 10, 2019
|
|
7W7V
 
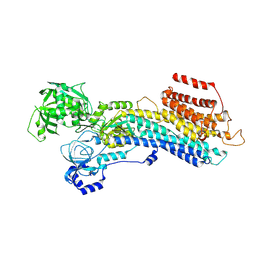 | | 'late' E2P of SERCA2b | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-14 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Multiple sub-state structures of SERCA2b reveal conformational overlap at transition steps during the catalytic cycle.
Cell Rep, 41, 2022
|
|
7W7U
 
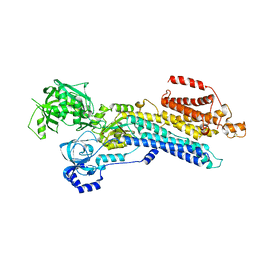 | | The 'Ca2+-unbound' BeF3- of SERCA2b | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-14 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Multiple sub-state structures of SERCA2b reveal conformational overlap at transition steps during the catalytic cycle.
Cell Rep, 41, 2022
|
|
