1M6N
 
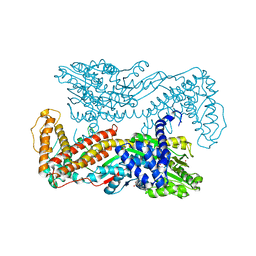 | | Crystal structure of the SecA translocation ATPase from Bacillus subtilis | | Descriptor: | Preprotein translocase secA, SULFATE ION | | Authors: | Hunt, J.F, Weinkauf, S, Henry, L, Fak, J.J, McNicholas, P, Oliver, D.B, Deisenhofer, J. | | Deposit date: | 2002-07-16 | | Release date: | 2002-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nucleotide Control of Interdomain Interactions in the Conformational Reaction Cycle of SecA
Science, 297, 2002
|
|
1M74
 
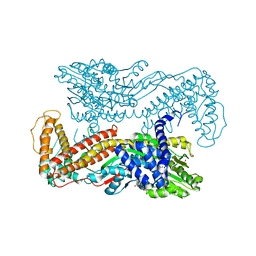 | | Crystal structure of Mg-ADP-bound SecA from Bacillus subtilis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Preprotein translocase secA, ... | | Authors: | Hunt, J.F, Weinkauf, S, Henry, L, Fak, J.J, McNicholas, P, Oliver, D.B, Deisenhofer, J. | | Deposit date: | 2002-07-16 | | Release date: | 2002-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Nucleotide Control of Interdomain Interactions in the Conformational Reaction Cycle of SecA
Science, 297, 2002
|
|
1G31
 
 | | GP31 CO-CHAPERONIN FROM BACTERIOPHAGE T4 | | Descriptor: | GP31, PHOSPHATE ION, POTASSIUM ION | | Authors: | Hunt, J.F, Van Der Vies, S.M, Henry, L, Deisenhofer, J. | | Deposit date: | 1998-03-27 | | Release date: | 1998-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural adaptations in the specialized bacteriophage T4 co-chaperonin Gp31 expand the size of the Anfinsen cage.
Cell(Cambridge,Mass.), 90, 1997
|
|
9NLB
 
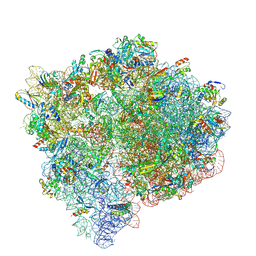 | |
9NLJ
 
 | | E.coli Initiation complex with Uup-EQ2 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Singh, S, Hunt, J.F. | | Deposit date: | 2025-03-03 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | E.coli Initiation complex with Uup-EQ2
To Be Published
|
|
9NKL
 
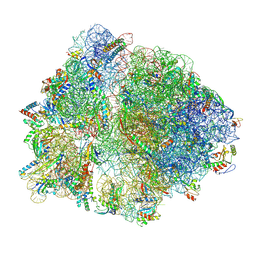 | | E. coli 70S initiation complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Singh, S, Hunt, J.F. | | Deposit date: | 2025-03-01 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | E. coli 70S initiation complex
To Be Published
|
|
9NLQ
 
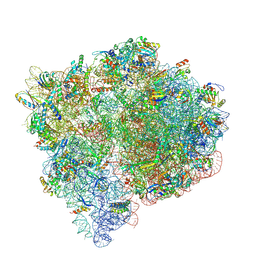 | |
9NJV
 
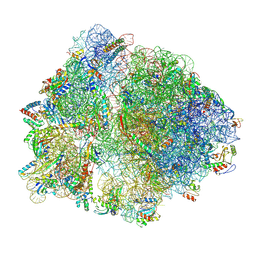 | |
9NLH
 
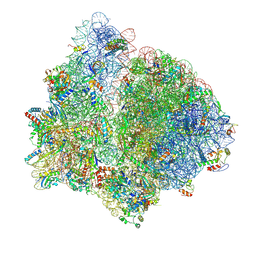 | | E.coli Initiation complex with YheS-EQ2 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Singh, S, Hunt, J.F. | | Deposit date: | 2025-03-03 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | E.coli Initiation complex with YheS-EQ2
To Be Published
|
|
9NL7
 
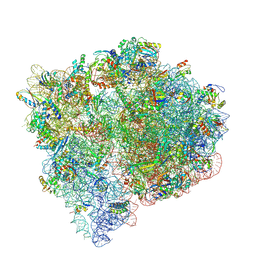 | |
9NLF
 
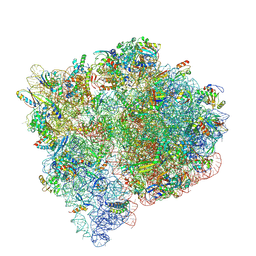 | |
9NLE
 
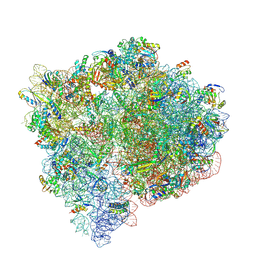 | |
3SM3
 
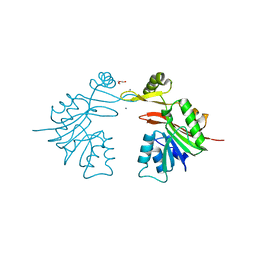 | | Crystal Structure of SAM-dependent methyltransferases Q8PUK2_METMA from Methanosarcina mazei. Northeast Structural Genomics Consortium Target MaR262. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, SAM-dependent methyltransferases | | Authors: | Vorobiev, S, Neely, H, Seetharaman, J, Snyder, A, Patel, P, Xiao, R, Ciccosanti, C, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-27 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Crystal Structure of SAM-dependent methyltransferases Q8PUK2_METMA from Methanosarcina mazei.
To be Published, 2009
|
|
4LNY
 
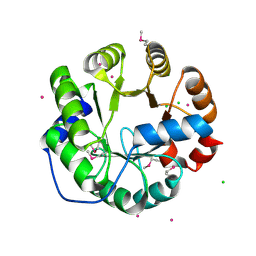 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR422 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Engineered Protein OR422 | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Sahdev, S, Xiao, R, Maglaqui, M, Kogan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Crystal Structure of Engineered Protein OR422.
To be Published
|
|
4LOA
 
 | | X-ray structure of the de-novo design amidase at the resolution 1.8A, Northeast Structural Genomics Consortium (NESG) Target OR398 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, De-novo design amidase | | Authors: | Kuzin, A, Lew, S, Vorobiev, S.M, Seetharaman, J, Sahdev, S, Xiao, R, Maglaqui, M, Kogan, S, Khersonsky, O, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR398
To be Published
|
|
1KXZ
 
 | | MT0146, the Precorrin-6y methyltransferase (CbiT) homolog from M. Thermoautotrophicum, P1 spacegroup | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, DeTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-01 | | Release date: | 2002-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of MT0146/CbiT Suggests that the Putative
Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1L3I
 
 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, ADOHCY BINARY COMPLEX | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-27 | | Release date: | 2002-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of Mt0146/CbiT Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
8GDY
 
 | | Crystal structure of the human PDI first domain with 9 mutations | | Descriptor: | 1,2-ETHANEDIOL, Protein disulfide-isomerase, THIOCYANATE ION | | Authors: | Forouhar, F, Banayan, N.E, Loughlin, B.L, Singh, S, Wong, V, Hunt, H.S, Handelman, S.K, Price, N, Hunt, J.F. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Systematic enhancement of protein crystallization efficiency by bulk lysine-to-arginine (KR) substitution.
Protein Sci., 33, 2024
|
|
8GDU
 
 | | Crystal structure of a mutant methyl transferase from Methanosarcina acetivorans, Northeast Structural Genomics Consortium (NESG) Target MvR53-11M | | Descriptor: | Methyltransferase domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Forouhar, F, Banayan, N.E, Loughlin, B.L, Singh, S, Wong, V, Hunt, H.S, Handelman, S.K, Price, N, Hunt, J.F. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Systematic enhancement of protein crystallization efficiency by bulk lysine-to-arginine (KR) substitution.
Protein Sci., 33, 2024
|
|
9NLS
 
 | |
9NJF
 
 | |
9NL5
 
 | |
9NL6
 
 | |
1TO0
 
 | | X-ray structure of Northeast Structural Genomics target protein sr145 from Bacillus subtilis | | Descriptor: | Hypothetical UPF0247 protein yyda | | Authors: | Kuzin, A.P, Edstrom, W, Vorobiev, S.M, Shastry, R, Ma, L.-C, Xiao, R, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-11 | | Release date: | 2004-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of Northeast Structural Genomics target protein sr145 from Bacillus subtilis
To be Published
|
|
1TZA
 
 | | X-ray structure of Northeast Structural Genomics Consortium target SoR45 | | Descriptor: | CACODYLATE ION, apaG protein | | Authors: | Kuzin, A.P, Vorobiev, S.M, Edstrom, W, Acton, T.B, Shastry, R, Ma, L.-C, Cooper, B, Xiao, R, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-07-09 | | Release date: | 2004-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of Northeast Structural Genomics Consortium
target SoR45
To be Published
|
|
