3CG7
 
 | |
3CM6
 
 | |
3CM5
 
 | |
3NH0
 
 | |
3NGZ
 
 | |
3NH2
 
 | |
3NGY
 
 | | Crystal structure of RNase T (E92G mutant) | | Descriptor: | COBALT (II) ION, Ribonuclease T, his tag sequence | | Authors: | Hsiao, Y.-Y, Yuan, H.S. | | Deposit date: | 2010-06-14 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural basis for RNA trimming by RNase T in stable RNA 3'-end maturation
Nat.Chem.Biol., 7, 2011
|
|
3NH1
 
 | |
3V9W
 
 | |
3V9Z
 
 | |
3V9S
 
 | |
3V9U
 
 | |
3VA3
 
 | |
3V9X
 
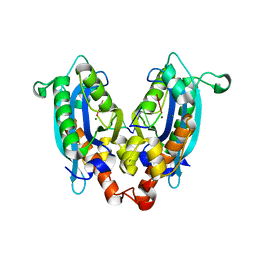 | |
3VA0
 
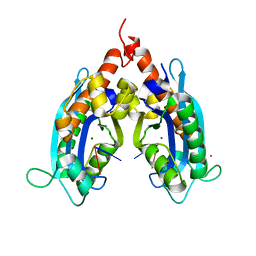 | |
5DK5
 
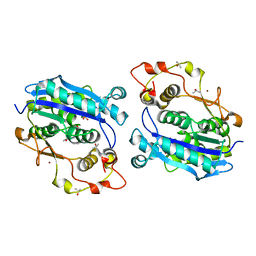 | | Crystal structure of CRN-4-MES complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cell death-related nuclease 4, ISOPROPYL ALCOHOL, ... | | Authors: | Hsiao, Y.-Y, Yuan, H.S. | | Deposit date: | 2015-09-03 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Inhibitors for the DEDDh Family of Exonucleases and a Unique Inhibition Mechanism by Crystal Structure Analysis of CRN-4 Bound with 2-Morpholin-4-ylethanesulfonate (MES)
J.Med.Chem., 59, 2016
|
|
1M07
 
 | | RESIDUES INVOLVED IN THE CATALYSIS AND BASE SPECIFICITY OF CYTOTOXIC RIBONUCLEASE FROM BULLFROG (RANA CATESBEIANA) | | Descriptor: | 5'-D(*AP*CP*GP*A)-3', Ribonuclease | | Authors: | Leu, Y.-J, Chern, S.-S, Wang, S.-C, Hsiao, Y.-Y, Amiraslanov, I, Liaw, Y.-C, Liao, Y.-D. | | Deposit date: | 2002-06-12 | | Release date: | 2003-01-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Residues involved in the catalysis, base specificity, and cytotoxicity of ribonuclease from Rana catesbeiana based upon mutagenesis and X-ray crystallography
J.Biol.Chem., 278, 2003
|
|
3HKM
 
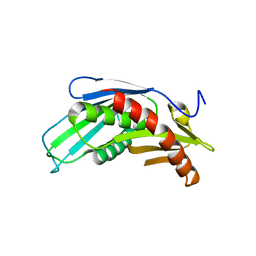 | | Crystal Structure of rice(Oryza sativa) Rrp46 | | Descriptor: | Os03g0854200 protein | | Authors: | Yang, C.-C, Wang, Y.-T, Hsiao, Y.-Y, Doudeva, L.G, Yuan, H.S. | | Deposit date: | 2009-05-25 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9845 Å) | | Cite: | Structural and biochemical characterization of CRN-5 and Rrp46: an exosome component participating in apoptotic DNA degradation
Rna, 16, 2010
|
|
3KRN
 
 | | Crystal Structure of C. elegans cell-death-related nuclease 5(CRN-5) | | Descriptor: | Protein C14A4.5, confirmed by transcript evidence | | Authors: | Yang, C.-C, Wang, Y.-T, Hsiao, Y.-Y, Doudeva, L.G, Chow, S.Y, Yuan, H.S. | | Deposit date: | 2009-11-19 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.918 Å) | | Cite: | Structural and biochemical characterization of CRN-5 and Rrp46: an exosome component participating in apoptotic DNA degradation
Rna, 16, 2010
|
|
4KAZ
 
 | | Crystal structure of RNase T in complex with a Y structured DNA | | Descriptor: | DNA (5'-D(*TP*TP*GP*GP*CP*CP*CP*TP*CP*TP*TP*TP*AP*GP*GP*GP*CP*CP*CP*C)-3'), MAGNESIUM ION, Ribonuclease T | | Authors: | Hsiao, Y.-Y, Yuan, H.S. | | Deposit date: | 2013-04-23 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into DNA repair by RNase T--an exonuclease processing 3' end of structured DNA in repair pathways.
Plos Biol., 12, 2014
|
|
4KB1
 
 | |
4KB0
 
 | |
5GYJ
 
 | | Structure of catalytically active sortase from Clostridium difficile | | Descriptor: | Putative peptidase C60B, sortase B | | Authors: | Yin, J.-C, Fei, C.-H, Hsiao, Y.-Y, Nix, J.C, Huang, I.-H, Wang, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural Insights into Substrate Recognition by Clostridium difficile Sortase.
Front Cell Infect Microbiol, 6, 2016
|
|
3DGT
 
 | | The 1.5 A crystal structure of endo-1,3-beta-glucanase from Streptomyces sioyaensis | | Descriptor: | Endo-1,3-beta-glucanase, MAGNESIUM ION | | Authors: | Li, T.H. | | Deposit date: | 2008-06-16 | | Release date: | 2008-09-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5 A structure of endo-1,3-beta-glucanase from Streptomyces sioyaensis: evolution of the active-site structure for 1,3-beta-glucan-binding specificity and hydrolysis
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4P5U
 
 | | Crystal structure of TatD | | Descriptor: | Tat-linked quality control protein TatD | | Authors: | Chen, Y, Li, C.-L, Hsiao, Y.-Y, Duh, Y, Yuan, H.S. | | Deposit date: | 2014-03-20 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of TatD exonuclease in DNA repair.
Nucleic Acids Res., 42, 2014
|
|
